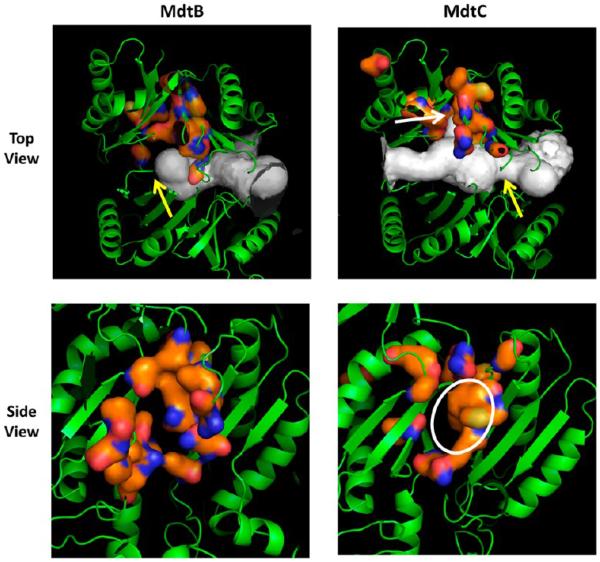
Figure 7.
Homology models of MdtB and MdtC. In the top panels, the models are shown in the orientation with the periplasmic, TolC-binding domains on top and the periplasmic cleft on the right, with the large tunnels detected by Caver colored white. The orange surface represents the putative deep binding pockets, corresponding to F136, V139, F178, I277, A278, E280, P285, Y327, F610, V612, F615, F617, I626, and F628 of AcrB.11 These are residues 146, 149, 187, 280, 282, 283, 288, 330, 606, 608, 611, 613, 622, and 624 in MdtB and residues 136, 139, 178, 271, 273, 274, 279, 321, 596, 598, 601, 608, and 610 in MdtC. In MdtB, the tunnel is closed in the middle (yellow arrow), whereas a constriction (yellow arrow) and a branch leading into the binding cavity (white arrow) are present in MdtC. In the bottom panels, the view is from the top (i.e., the TolC-binding domain). The putative substrate-binding pocket is relatively narrow in MdtB, whereas there is a substantial hydrophobic area in the wide pocket of MdtC (white ellipse). In both panels, the figures show clipped views so that the binding sites can be seen easily. They were produced by using PyMol (http://www.pymol.com).