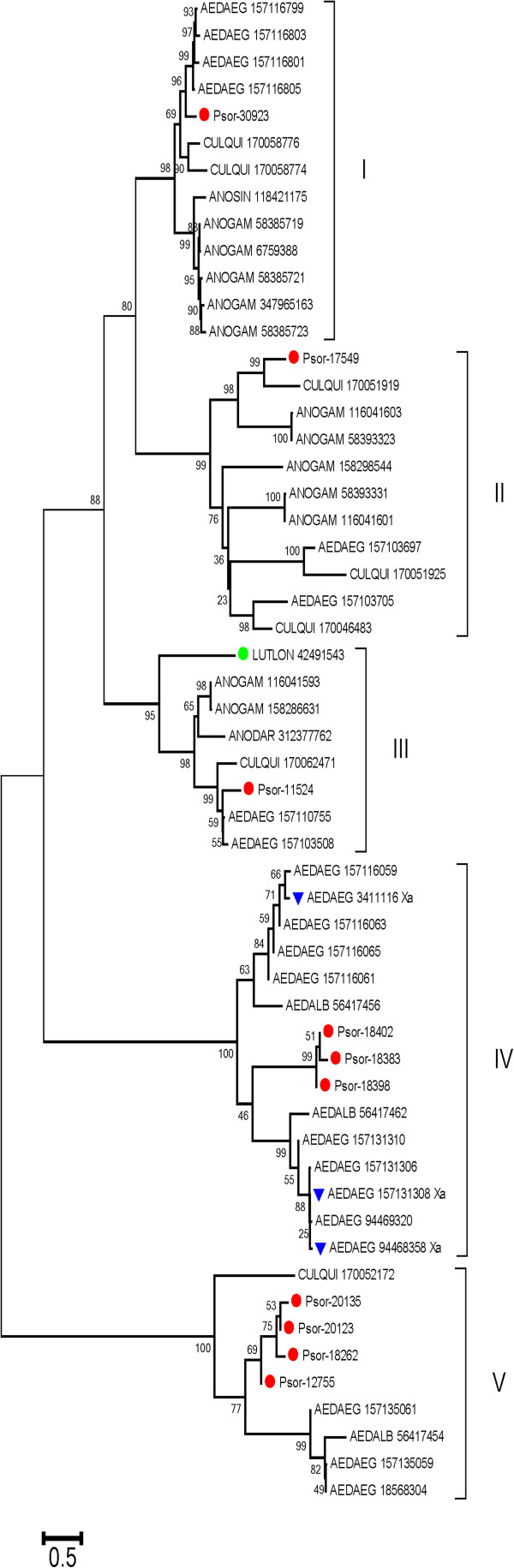
Figure 2.
Phylogram of the salivary serpins of Psorophora albipes. The bootstrapped (1,000 iterations) phylogram was obtained using the serpins from P. albipes and their best-matching proteins from the non-redundant database from NCBI. P. albipes proteins are recognized by the prefix Psor- and a red marker (red circle symbol). Other proteins are represented by the first three letters of the genus name, followed by the first three letters of the species name, followed by their gi| accession number. The Aedes aegypti proteins identified as anti Xa anticlotting are marked with a blue symbol (blue reversed triangle symbol). The sole Lutzomyia longipalpis protein is identified by a green symbol (green circle symbol).The numbers at the nodes represent bootstrap support for 1,000 iterations using the neighbor-joining algorithm. The bar at the bottom indicates 50% amino acid substitution. Roman numerals indicate individual clades with strong bootstrap support.