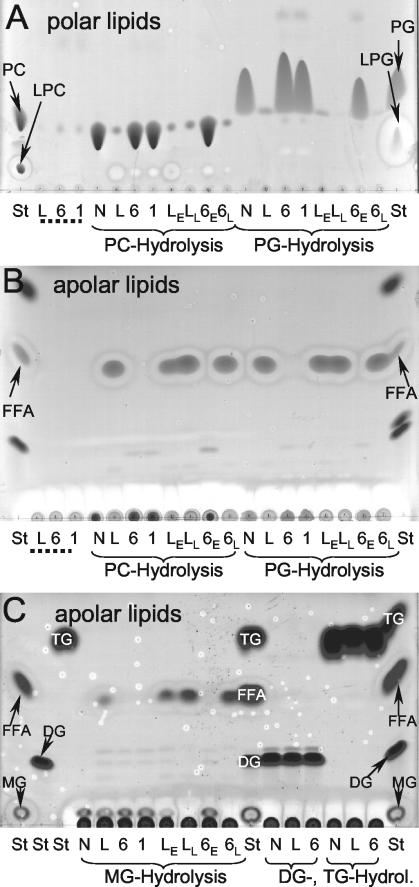
FIG. 5.
TLC analysis of lipid hydrolysis by cell lysates of wild-type, plaB mutant, and genetically complemented L. pneumophila strains. Cell lysates of wild-type L. pneumophila (L), plaB mutants plaB60 (6) and plaB1 (1), or genetically complemented L. pneumophila (wild type and plaB60 mutant harboring the empty vector [LE and 6E, respectively] and wild type and plaB60 mutant harboring pKH192, containing intact plaB [LL and 6L, respectively]) were incubated with DPPC (PC) or DPPG (PG) (A and B) or with 1-MPG (MG), 1,2-DG (DG), or TG (C) for 24 h at 37°C, and then lipids were extracted and subjected to TLC. A polar or apolar solvent mixture was used for the separation of the polar (A) or apolar (B and C) lipids, respectively. A mixture of Tris-HCl buffer and the lipids was also incubated and served as a negative control (N). Furthermore, bacterial-cell lysates not incubated with any lipid were extracted and served as the bacterial lipid background control (designated by dotted lines). In all incubations, samples were examined for degradation of the lipid substrate (for DPPC and DPPG, polar TLC [A]; for 1-MPG, 1,2-DG, and TG, apolar TLC [C]), enrichment of FFA (apolar TLC [B and C]), and in the case of DPPC and DPPG, incubations for enrichment of the respective lysophospholipid (polar TLC [A]). For qualitative identification of the lipid spots, lanes containing lipid standards (St) were included. The observations depicted here were made on one more occasion.