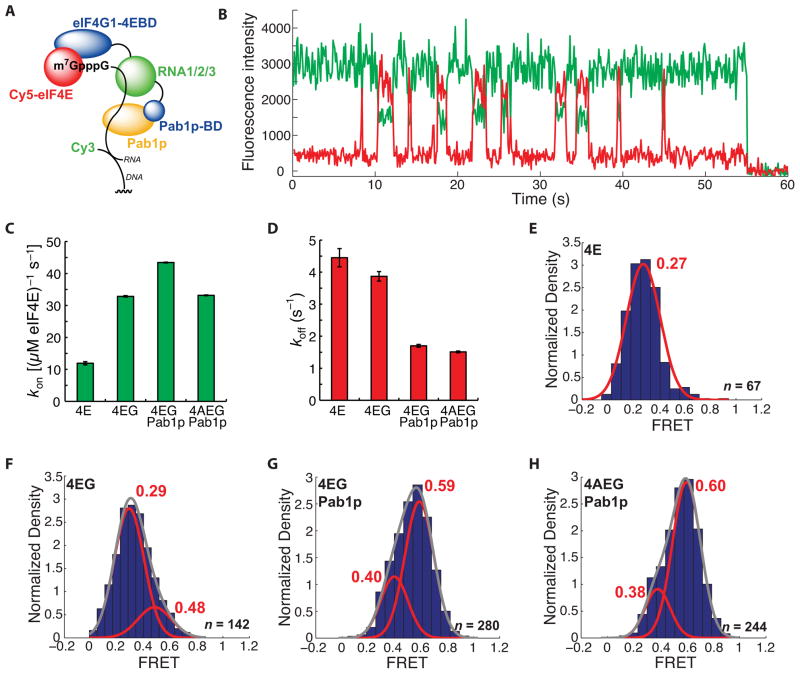
Figure 4.
A long-lived, high-FRET state for the Cy5-eIF4E•RNA complex observed in the presence of Pab1p. (A) Schematic of experimental design. The domain architecture of eIF4G1 is abridged from the full disposition of domains (Figure 2B) by combining the three separate RNA binding domains and omitting the eIF4A-binding domain. Pab1p-bd refers to the eIF4G1 Pab1p-binding domain. (B) Representative single-molecule FRET trace showing binding of Cy5-eIF4E to immobilized, cap-poly(CU) RNA in the presence of 2 μM Pab1p. (C) Dependence of Cy5-eIF4E-mRNA association rate on factor composition. (D) Dependence of dissociation rate of the Cy5-eIF4E•mRNA complex in the presence of eIF4G1 (250 nM), Pab1p (2 μM), and eIF4A (400 nM). (E) – (H) FRET intensity distributions for the Cy5-eIF4E complexes in (D). The number of single-molecule traces (n) used to construct distributions is given where appropriate. Error bars in kinetic plots represent standard errors for single-exponential fits. See also Figure S4.