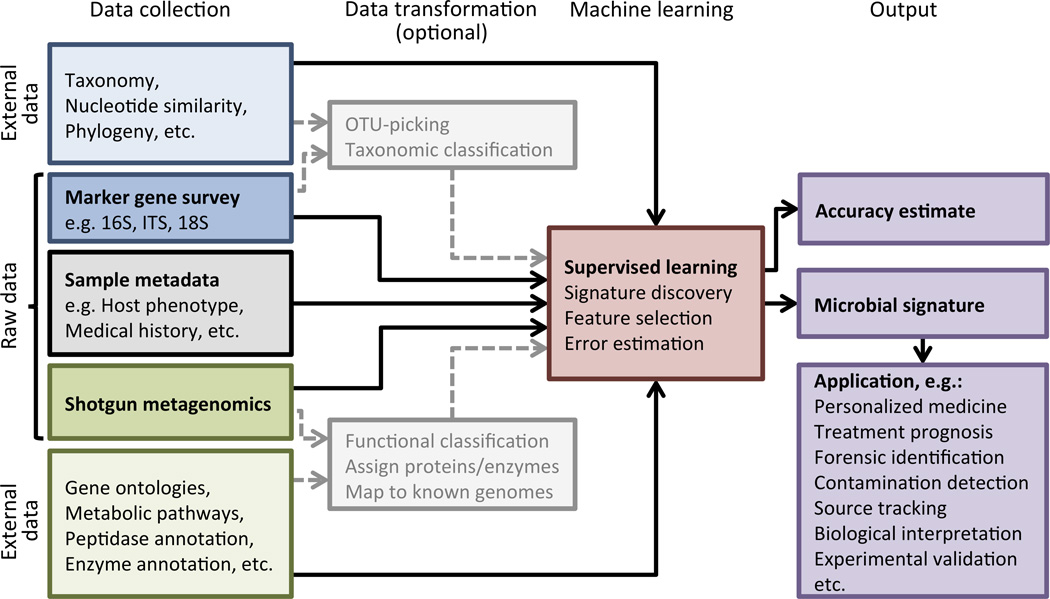
Figure 1. Processes for microbial signature discovery.
The process begins with the collection of a large set of sequencing data from various bacterial communities associated with different environments or different host phenotypes. These sequences can serve directly as input to a machine learning algorithm, or they can be transformed through a preprocessing step (data transformation). Although for microbial community analysis data transformation and supervised learning are typically performed as separate steps, we suggest that predictive models will be improved by the development of novel machine learning techniques that are informed by the potential data transformations. For example, constructing a good predictive model using metabolic characterizations of metagenomics sequences might be easier if the algorithm has knowledge of the hierarchical relationships between metabolic functions. In the case of marker-gene surveys, a machine learning algorithm may benefit from knowledge of the phylogenetic relationships of the observed lineages, or the network of average nucleotide similarities between the input sequences. These structures may allow models to share statistical strength across related independent variables in cases where there is high variability within a given environment or host phenotype (i.e. lack of a “core microbiome”).