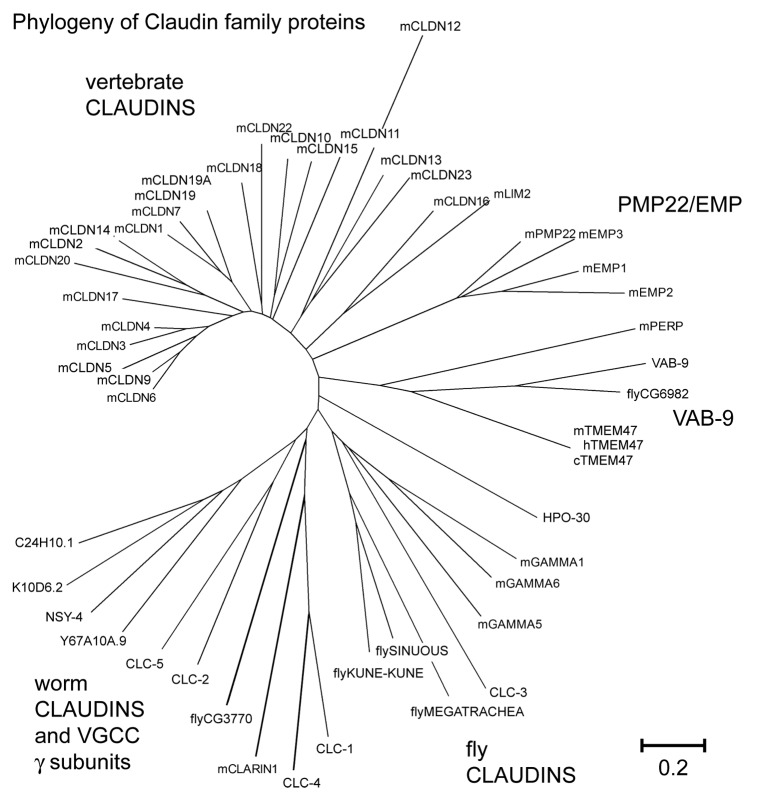
Figure 2. pfam00822 proteins from vertebrates and invertebrates are highly divergent. Bootstrap analysis of pfam00822 proteins using the MEGA (Molecular Evolutionary Genetic Analysis) software program available at http://www.megasoftware.net.108 The phylogenetic tree resulting from ClustalW2 analysis is shown. Several different subgroup clades are observed. Note that NSY-4 clusters with other putative gamma subunits and C. elegans claudins, while the vertebrate VGCC gamma subunits are loosely associated with fly claudins. Scale bar indicates amino acid substitutions per residue. Accession numbers for proteins included in the alignments are: NSY-4 (NP_500189.4), K10D6.2(NP_505843), R04F11.1 (NP_506087), C24H10.1(NP_508863), Y67A10A.9(NP_502746.1), mGamma5 (voltage-dependent calcium channel gamma subunit 5) (NP_542375), mGamma1 (NP_031608), mGamma6 (NP_573446.1), mGamma3 (NP_062303,)CLC-1(NP_509847), CLC-2(NP_509257), CLC-3 isoform a(NP_001024993), CLC-4 (NP_509800), CLC-5(NP_509258), VAB-9 (NP_495836), CG6982 (dVAB-9) (NP_001097876), mTMEM47(NP_620090), cTMEM47 (NP_001003045.1), hTm47 (NP_113630.1), xtmem47 (NP_001085134.1), mclaudin1 (NP_057883), mclaudin2 (NP_057884), mclaudin3 (NP_034032), mclaudin4 (NP_034033), mclaudin5 (NP_038833), mclaudin6 (NP_0247), mclaudin7 (NP_058583), mclaudin9 (NP_064689), mclaudin10 (a) (NP_076367), mclaudin11 (NP_032796), mclaudin12 (NP_075028), mclaudin13 (NP_065250), mclaudin14 (NP_001159398), mclaudin15 (NP_068365), mclaudin16 (NP_444471), claudin17 (NP_852467), mclaudin18(NP_062789), mclaudin19(1)(NP_001033679), Claudin19(2)(NP_694745), mclaudin20 (NP_001095030), mclaudin22 (NP_083659), mclaudin23 (NP_082274), fly_Sinuous (a) (NP_647971), fly_Megatrachea (NP_726742), fly_CG3770 (NP_611985.), fly_Kune-kune (NP_610179), mEMP2 (NP_031955), mPMP22 (NP_032911), mEMP3(NP_001139818), mEMP1(NP_034258), mPERP(NP_071315), LIM2 (NP_808361)

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.