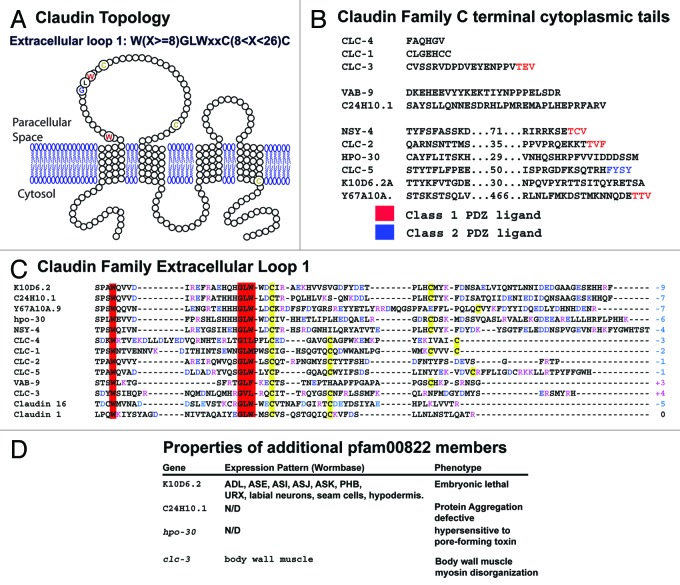
Figure 3. Sequence conservation and motifs in C. elegans pfam00822 proteins. (A) The predicted membrane spanning topology of VAB-9 is shown; pfam00822 proteins have the same membrane-spanning topology. Highly conserved residues among family members are indicated in enlarged, colored circles. (B) The C-terminal cytoplasmic tails of the C. elegans pfam00822 proteins are shown, from the end of the final transmembrane domain to the final residue. Sequences of longer proteins are abbreviated; numbers indicate intervening residues not shown. Terminal sequences conforming to class 1 and 2 PDZ binding domain ligand rules are indicated in red and blue, respectively. (C) The first extracellular loop domain of C. elegans proteins and murine claudin are shown. Postiviely charged residues are indicated in red, negative charged residues in blue. The sum of extracellular loop charge is indicated at the end of the sequence. Cysteines are indicated in yellow and the conserved tryptophan and GLW motif is indicated in red. (D) Additional pfam00822 proteins, for which some preliminary information is known, are shown. K10D6.2 expression, C24H10 function in protein aggregation, hpo-30 function in toxin resistance, and clc-3 expression and phenotype are referenced in 109, 110, 111, 112, and 113 respectively. Additional unpublished results, particularly regarding clc-3, can be found at http://wormbase.org.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.