Fig. 3.
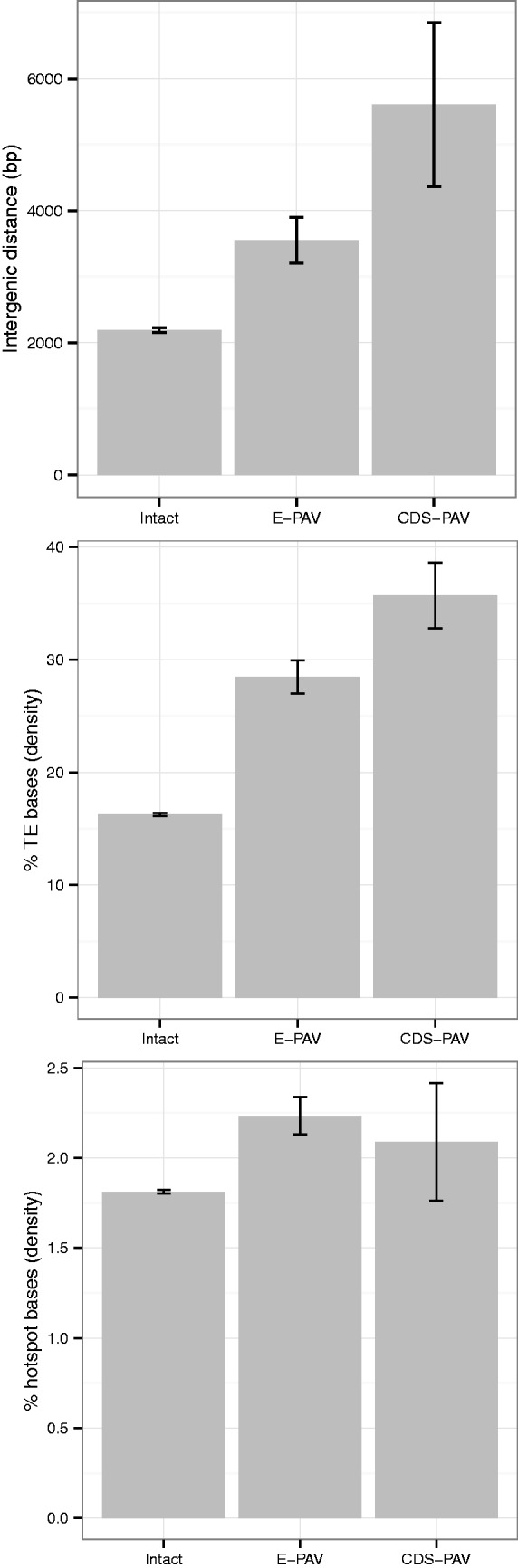
Genomic context for intact (having no exons under P/A variation), E-PAV (having at least one, but not all, exons missing in at least one accession), and CDS-PAV (having the entire CDS missing in at least one accession) genes. Averaged values for the genes in each set are given for, from top to bottom, the intergenic distance, the percentage of TE bases in the nongenic sequence of a 10-kb window centered on that gene’s midpoint, and the percentage of recombinogenic motifs in the genic sequence of a 1-kb window centered on that gene’s midpoint. See also supplementary tables 3 and 4 (Supplementary Material online) for the values of specific TE families and other window sizes.
