Abstract
Animal venoms have evolved many times. Venomous species are especially common in three of the four main groups of arthropods (Chelicerata, Myriapoda, and Hexapoda), which together represent tens of thousands of species of venomous spiders, scorpions, centipedes, and hymenopterans. Surprisingly, despite their great diversity of body plans, there is no unambiguous evidence that any crustacean is venomous. We provide the first conclusive evidence that the aquatic, blind, and cave-dwelling remipede crustaceans are venomous and that venoms evolved in all four major arthropod groups. We produced a three-dimensional reconstruction of the venom delivery apparatus of the remipede Speleonectes tulumensis, showing that remipedes can inject venom in a controlled manner. A transcriptomic profile of its venom glands shows that they express a unique cocktail of transcripts coding for known venom toxins, including a diversity of enzymes and a probable paralytic neurotoxin very similar to one described from spider venom. We screened a transcriptomic library obtained from whole animals and identified a nontoxin paralog of the remipede neurotoxin that is not expressed in the venom glands. This allowed us to reconstruct its probable evolutionary origin and underlines the importance of incorporating data derived from nonvenom gland tissue to elucidate the evolution of candidate venom proteins. This first glimpse into the venom of a crustacean and primitively aquatic arthropod reveals conspicuous differences from the venoms of other predatory arthropods such as centipedes, scorpions, and spiders and contributes valuable information for ultimately disentangling the many factors shaping the biology and evolution of venoms and venomous species.
Keywords: venomics, venom evolution, agatoxin, ESTs, venomous arthropods
Introduction
Venoms have evolved many times in the animal kingdom, and they play important roles in predation, defense, competition, communication, and defense against harmful microorganisms (Fry et al. 2009; Hölldobler and Wilson 2009; dos Santos Pinto et al. 2012; Casewell et al. 2013). Venoms are generally distinguished from poisons as being mostly proteinaceous secretory products synthesized in specialized glands and by their active mode of delivery through wounds inflicted by specialized structures (Fry et al. 2009; Casewell et al. 2013). The specific activities of a venom are largely determined by the specific mix of its peptide and protein components, which are individually referred to as toxins. The study of venom composition, which is greatly assisted by recent advances in transcriptomic and proteomic technologies (Casewell et al. 2013), is therefore a critical first step toward understanding a venom’s biological role and potential applied uses (King 2011). The study of venom toxins has also proven valuable for generating insights into fundamental biological questions, such as the role of gene duplication in producing evolutionary novelties, the processes involved in generating large multigene families, and the molecular bases of ecological adaptations (Casewell et al. 2011; Sunagar et al. 2012; Brust et al. 2013). However, general insights are only possible insofar as we understand taxon-specific aspects of venom biology and evolution. Hence, it is important to expand our understanding of the diversity of venoms beyond the best studied taxa (principally snakes, spiders, scorpions, and cone snails).
Arthropoda is a clade especially rich in venomous species. Three of the four main groups (Chelicerata, Hexapoda, and Myriapoda) together represent tens of thousands of venomous animals inhabiting a broad range of mostly terrestrial ecosystems. However, despite their unparalleled diversity of body plans (Martin and Davis 2001), no unambiguous evidence exists that any crustacean is venomous.
Remipedes were described in 1981 (Yager 1981) and are blind crustaceans that inhabit anchialine underwater cave systems (Koenemann et al. 2007; von Reumont and Burmester 2010). The older perspective that the remipedes have one of the most primitive crustacean body plans (Schram and Hof 1998), with a long homonomously segmented trunk with similar biramous swimming limbs, has been challenged in recent studies, which instead suggest that remipedes are more derived crustaceans that are closely related to hexapods (Ertas et al. 2009) within a clade Pancrustacea (crustaceans+hexapods) (Fanenbruck et al. 2004; Ertas et al. 2009; Regier et al. 2010; von Reumont et al. 2012). Intriguingly, the first phylogenomic study that included data on remipedes (von Reumont et al. 2012), strongly supports the hypothesis that remipedes are the closest crustacean relatives of hexapods. It has been suspected, mainly based on morphology, that remipedes are venomous (van der Ham and Felgenhauer 2007a, 2007b), but in the absence of data on the composition of their putative venom, their status as venomous organisms is tentative.
We present in this study the first conclusive evidence that remipedes are in fact venomous crustaceans. Transcriptomic profiling of toxin gene expression shows that the remipede’s venom glands express a unique mix of known venom toxins. We supplement these data by the most detailed 3D reconstruction of the functional morphology of the remipede venom delivery apparatus produced to date to show that the animals can inject venom in a controlled manner. Taking into account the recent insight (Casewell et al. 2012) that studies of venom toxin evolution can be biased by restricting analyses to venom gland tissue, we used a whole animal transcriptome to identify nontoxin paralogs, which allowed us to reconstruct the probable evolutionary origin of a unique remipede neurotoxin.
Results and Discussion
Remipedes Inject Their Venom via a Highly Developed Venom Delivery Apparatus
Remipedes have a superficially centipede-like habitus, with long homonomously segmented bodies, with most segments equipped with biramous swimming legs. The head, or cephalon, in contrast, features a robust pair of legs (maxillules) that in most species end in a sharp tip.
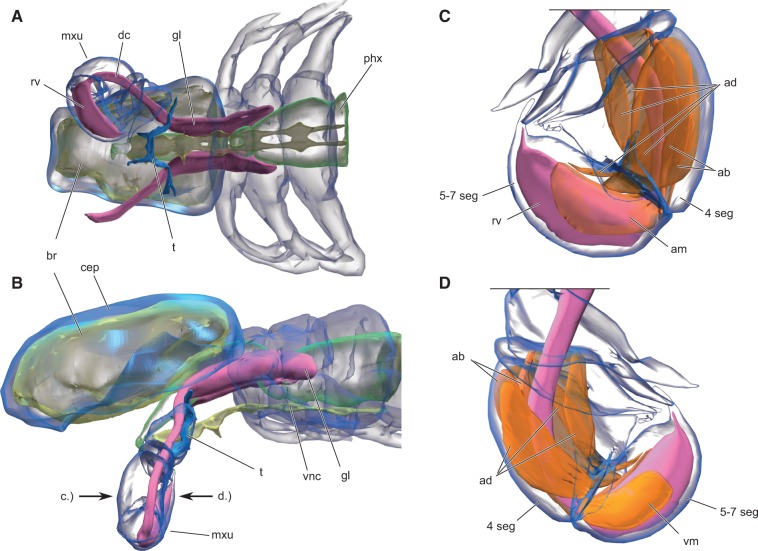
We used synchrotron radiation micro-computer tomography (SR-µCT) to prepare the first three-dimensional reconstruction of the venom delivery apparatus of the remipede Speleonectes tulumensis. Our results show a highly adapted venom delivery apparatus. The anterior trunk of S. tulumensis contains two equally sized venom glands, which connect via ducts to reservoirs located in the terminal segments of the maxillules (fig. 1). Two large muscles (am and vm in fig. 1c and d) closely oppose the anterior and posterior sides of the reservoir. They insert only at the mediodistal part of the subterminal segment of the maxillule, which indicates that they cannot function in limb movement, but that contraction of these muscles will expel venom from the reservoir. The stabbing motion of the maxillule, which remipedes employ on prey (Carpenter 1999), results from the contraction of four adductor muscles located in the subterminal segment, which at the same time passively prevents backflow of the venom by compressing the duct. This muscle arrangement in S. tulumensis clearly represents an effective venom delivery system. These findings expand our knowledge of the functional morphology of remipede maxillules (van der Ham and Felgenhauer 2007a) by showing that limb movement and venom injection can be functionally separated.
Fig. 1.
Three-dimensional reconstructions of Speleonectes tulumensis (Crustacea: Remipedia) from high-resolution SR-µCT data. (A) Ventral and (B) lateral view showing the course of the venom delivery system (VDS), (purple) and its position inside the body. (C) Anterior and (D) posterior view focused on the maxillule and the muscle equipment related to the VDS. 4 seg, 4th segment; 5-7 seg, 5–7th segment; ab, abductors; ad, adductors; am, anterior apodemal muscle; br, brain; cep, cephalon; dc, ductus; gl, gland; mxu, maxillule; phx, pharynx; rv, reservoir; t, tentorium; vm, ventral apodemal muscle; vnc, ventral nerve cord. Images not to scale to each other, mouth parts and other structures are not shown.
Composition and Molecular Evolution of Remipede Venom
Our next-generation sequencing-based transcriptomic analysis revealed that 1,052 unique contigs were expressed in the venom glands of S. tulumensis. Of these, 191 are secretory proteins and 108 can be assigned to known venom toxin classes (supplementary figs. S1, S2a, and S2b, Supplementary Material online). For all the 191 proteins, InterProscan results were found, except for one (the lowly expressed isotig00521, see supplementary table S1, Supplementary Material online). Only six contigs received no Blast-hits at all, although they did receive InterProscan data results. Interestingly, the remipede’s neurotoxin sequences were among these (see supplementary table S1, Supplementary Material online). This small number of unidentifiable transcripts is in striking contrast to what is found in other venomous arthropods, such as scorpions, centipedes, and parasitic wasps. In these taxa, more than a third to more than half of all venom gland transcripts are unique and unidentified (Vincent et al. 2010; Morgenstern et al. 2011; Liu et al. 2012).
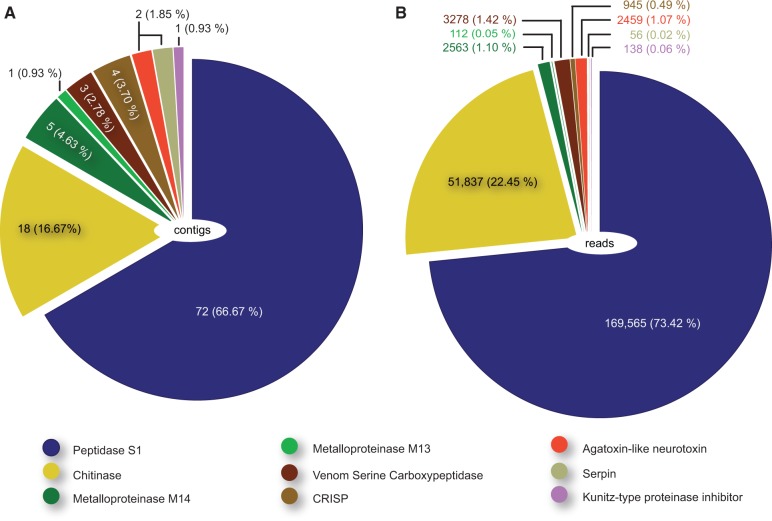
The three most diverse and abundant types of toxins expressed by the venom glands are proteinases, chitinases, and a putative neurotoxin, which together comprise more than 90% of toxin transcript diversity and more than 95% of toxin sequence reads (fig. 2). It should, of course, be noted that in the absence of functional data about proteins, our results can only designate sequences as putative toxins. This caveat should be kept in mind when interpreting our results. Given that remipedes are very difficult to collect and occur in small populations, it will be challenging to perform the proteomic work needed to confirm the presence and activities of the identified putative toxins in secreted remipede venom.
Fig. 2.
Transcriptomic profile of toxin genes expressed in the remipede venom glands. (A) Distribution of the diversity of contigs for each toxin type. (B) Distribution of the abundance of sequence reads per toxin type. Total numbers are given followed by percentages shown in brackets. The color code is given in the figure. See supplementary figures S2a and S2b, Supplementary Material online, for transcriptomic profiles of all secreted proteins expressed in both venom gland and complete animal libraries.
Peptidase S1
The peptidase S1 (PS1) family of serine proteases is a large peptidase family, and its members have many different biological roles, ranging from digestion to immune responses. Proteins of this gene family have been independently recruited into the venoms of several taxa, including cephalopods, hymenopterans, reptiles, and mammals (Aminetzach et al. 2009; Fry et al. 2009; Whittington et al. 2010; Ruder et al. 2013). In some of these, PS1s are both diverse and abundantly expressed, such as in the posterior venom glands of several species of cephalopods and the crural glands of male platypuses. Depending on the type, venom serine proteases have a variety of activities, such as the prevention of blood coagulation, and the causing of vasodilation, smooth muscle contraction, pain, and inflammation. PS1s represent the dominant component of the remipede’s venom gland transcripts in terms of sheer diversity and coverage (fig. 2). More than 66% of the venom gland toxin transcripts and more than 73% of venom gland toxin reads (in 37 isogroups) are PS1. Virtually all the venom gland PS1s contain the catalytic triad of His-Asp-Ser that is characteristic of this peptidase family (positions 69, 160, and 458; see supplementary fig. S3a, Supplementary Material online, and alignment data), and 10 conserved cysteine residues. The whole animal library also shows the expression of a large diversity of PS1s. However, the majority of the venom gland PS1s are restricted to a single clade of remipede-specific PS1s (supplementary fig. S3b, Supplementary Material online). Although comparable quantitative data derived from deep transcriptomic sequencing are not yet available for many taxa, the extraordinary diversity and level of expression of PS1 found in remipede venom glands are to our knowledge unique in the animal kingdom. Transcriptomic analyses of scorpion, spider, centipede, and hymenopteran venom glands show that serine peptidases as a whole account for only a small proportion of toxin transcripts (Vincent et al. 2010; Morgenstern et al. 2011; Almeida et al. 2012; Liu et al. 2012).
Several of the remipede PS1 sequences contain short insertions within the region encompassed by the catalytic triad. Interestingly, insertions found in this region of kallikrein-type PS1s expressed in the salivary glands of the shrew Blarina brevicauda have been hypothesized to be associated with possibly enhancing enzymatic activity (Aminetzach et al. 2009). A similar insertion reported from a kallikrein toxin expressed in the venom glands of the Gila monster (Heloderma horridum), however, was subsequently found to be a sequencing artifact (Fry et al. 2010). The high expression of the diverse remipede PS1s suggests that proteolytic activities play an important role in remipede envenomation, possibly aiding both prey immobilization and digestion. The latter role seems particularly relevant given that in one of the few field observations of remipede feeding they were observed to ingest the internal tissue of a prey crustacean, leaving behind a hollowed out cuticle (Schram and Lewis 1989).
Chitinases
Chitinases are the second most diverse and abundant toxin type expressed in the venom glands of S. tulumensis, representing more than 16% of toxin contigs, and more than 22% of toxin reads. Chitinases are a widely distributed group of hydrolytic enzymes that catalyze the breakdown of chitin, and they have so far been found to be expressed in the venom glands of octopodiform cephalopods, sicariid and theraphosid spiders, parasitoid hymenopterans, the secretions of hematophagous soft ticks, and hydrozoan nematocysts (Chen et al. 2008; Fernandes-Pedrosa et al. 2008; Fry et al. 2009; Vincent et al. 2010; Balasubramanian et al. 2012; Ruder et al. 2013) but never as such a major component as found in the remipede. By way of comparison, although chitinases are classified among “high abundance transcripts” in the venom of the sicariid spider Loxosceles laeta (Fernandes-Pedrosa et al. 2008), they account for less than 3% of known or possible toxins found in its venom. Similarly, chitinases account for less than 5% of toxin expressed sequence tags expressed in the venom gland of the parasitoid wasp Chelonus inanitus (Vincent et al. 2010). No chitinases are known to be expressed in the venom glands of centipedes and scorpions.
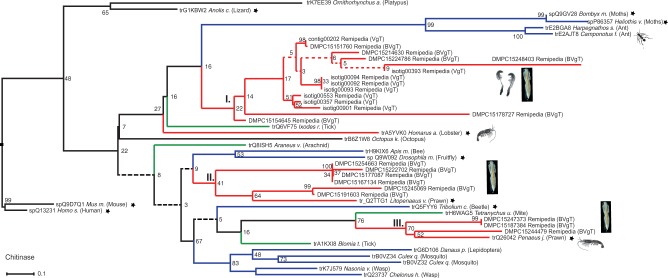
As can be seen in supplementary figure S2b, Supplementary Material online, the diversity of chitinase transcripts represents a substantially higher proportion of secreted and putative toxin genes in the venom gland library than in the complete animal library. Phylogenetic analysis reveals that the remipede chitinases are distributed across three putative clades. The chitinases expressed in the venom gland library are restricted to one remipede-specific clade (clade I in fig. 3). The two other clades only contain sequences derived from the whole animal library. The conservation of all nine active sites (alignment positions 133–141 in fig. 4) in remipede venom gland chitinases suggests that remipede venom has substantial chitinase activity. We speculate that this assists remipedes in softening and breaking down the chitinous exoskeletons of crustacean prey, especially the internal apodemes that provide anchors for their soft tissues. This hypothesis is consistent with available field observations, which suggest that crustaceans are probably an important component of the remipede diet (Carpenter 1999; Koenemann et al. 2007; van der Ham and Felgenhauer 2007b).
Fig. 3.
Phylogenetic tree of the chitinase sequences. The tree is reconstructed with RAxML (Stamatakis and Alachiotis 2010), -f a, PROTGAMMAWAG, 10,000 bootstraps. Nodes supported below 10% bootstrap values are represented by dashed lines. Chelicerates are colored in green, centipedes in brown, insects in blue, and crustaceans in red. Nonvenomous and nonhematophagous taxa are indicated by asterisks. Clade numbers are given in roman numerals. Venom gland transcripts and complete animal tissue transcripts are indicated by the abbreviations VgT and BVgT in parentheses following the sequence name, respectively. Remipede sequences are illustrated by miniaturized remipede pictures, and venom gland pictograms highlight remipede venom gland sequences.
Fig. 4.
Fraction of aligned domain region of chitinase sequences featuring the active sites. Excluded partitions of the alignment are indicated by brackets. The domain region is shaded in green and the active sites in red. For remaining description and color code see figure 3. Venom gland transcripts and complete animal tissue transcripts are indicated by the abbreviations VgT and BVgT in parentheses following the sequence name, respectively.
Remipede Venom Neurotoxin
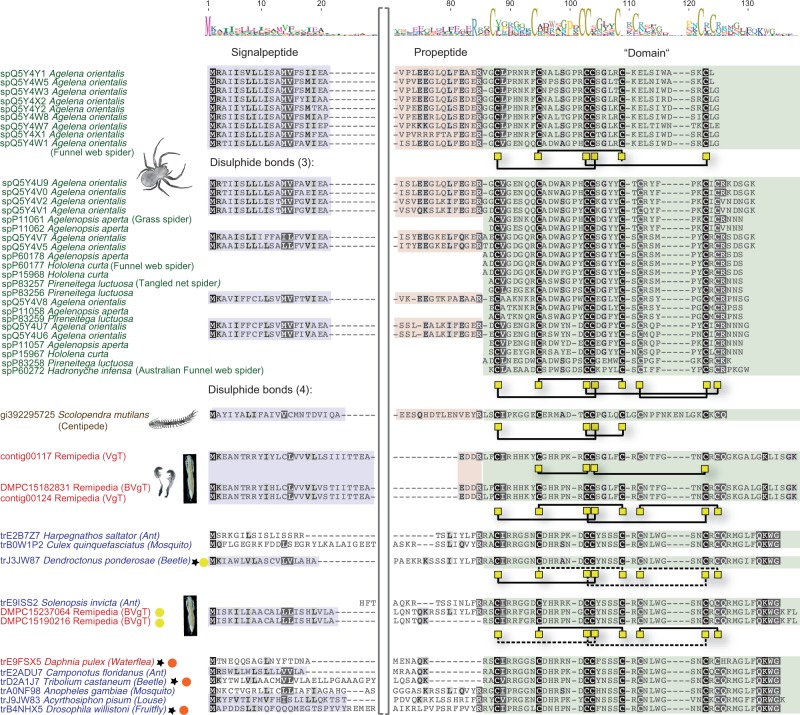
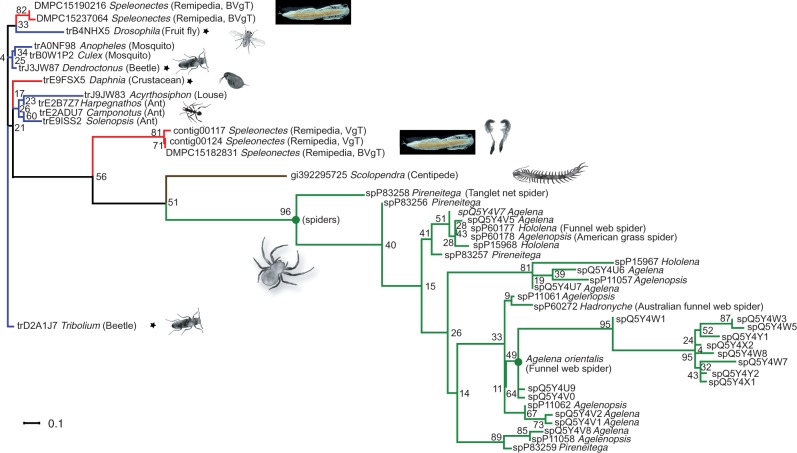
The enzymatic activity of the peptidases and chitinases probably plays a significant role in prey liquefaction. However, they are unlikely to cause rapid immobilization of prey. This important role could be fulfilled by the fifth most highly expressed remipede venom gland toxin. This toxin is represented by two distinct contigs that have the conserved cysteine pattern characteristic of ß/∂ agatoxins with virtually identical spacing [C-x(6)-C-x(6)-C-C-x(4)-C-x-C-x(6)-C-x-C] (fig. 5). ß/∂ agatoxins are a recently described type of spider venom neurotoxin (Billen et al. 2010), which causes presynaptic voltage-gated sodium channels to open at resting membrane potentials in insects. The resulting neurotransmitter release generates a stream of action potentials in motor neurons, resulting in irreversible spastic paralysis of the victim. Prediction of the presence of disulphide bonds suggests that the two remipede venom gland neurotoxin sequences are able to adopt two different types of folding, one with two disulphide bonds and one with four as in spider ß/∂ agatoxins (fig. 5).
Fig. 5.
Alignment of the remipede neurotoxin. The color code is identical to that in figure 3. Signal peptides are shaded in blue, propeptides in red, and the domain region in green. Disulphide bonds are pictured with black lines and the cysteine residue backbone by yellow squares. Red circles represent sequences of nonvenomous taxa that lack disulphide bonds as predicted using DBCP webserver (Lin and Tseng 2010). Yellow circles show nonvenomous taxa and remipede sequences that are predicted to have one to three less likely (<65% of probability) disulphide bonds, and the dotted black lines indicate these less likely bonds. Venom gland transcripts and complete animal tissue transcripts are indicated by the abbreviations VgT and BVgT in parentheses following the sequence name, respectively.
The whole animal library also expresses a putative neurotoxin sequence that is virtually identical to one of the venom gland sequences. We suspect that this sequence derives from the venom gland tissue that was part of the whole animals from which this library was prepared. Interestingly, the complete animal library also yielded two different sequences with strong sequence similarities to ß/∂ agatoxins, conserving the same pattern and spacing of cysteine residues, and with the predicted ability to form disulphide bonds. These sequences, which are probably expressed in the nonvenom gland tissue that dominates the complete animal library, differ from those found expressed in the remipede venom glands by having distinct signal peptides and a larger putative propeptide region. Intriguingly, a Blast search yielded a number of pancrustacean (several hexapod species and the crustacean Daphnia) sequences with an identical cysteine pattern and spacing as the putative remipede neurotoxin sequences. These sequences are remarkably similar to the remipede sequences obtained from the complete animal library, with strong sequence similarities in the mature chain and with most possessing a similarly large putative propeptide (fig. 5).
The results of our phylogenetic analysis (fig. 6 and supplementary fig. S4, Supplementary Material online) show an intriguing pattern. To our knowledge they show for the first time that spider agatoxins can be linked to homologs in nonspiders. The gene tree that includes sequences both from venomous and nonvenomous arthropods shows that the putative remipede venom gland neurotoxin is clearly separated on a long branch from its nonvenom gland paralog, which instead is part of a short-branch cluster comprising nonvenom-gland-derived sequences from other crustaceans and insects. This separation of the putative remipede venom gland neurotoxin from the remaining pancrustacean sequences is independent of two possible positions of the root of the tree. One rooting position is between the pancrustaceans and the centipede plus spiders, reflecting the consensus topology of higher level arthropod phylogeny (Giribet and Edgecombe 2012; von Reumont et al. 2012), whereas the alternative, which we have chosen, is rooting the tree with one of the nonvenom gland pancrustacean sequences, such as that from Tribolium. In both cases, our results are consistent with the hypothesis that the remipede venom gland neurotoxin represents a paralog of that expressed in the nonvenom gland tissue (see also supplementary fig. S4, Supplementary Material online). This suggests that the origin of the remipede venom gland neurotoxin is associated with substantial changes in all regions of the primary sequence of the protein, with probable consequences for the level and location of gene expression as well as the folding and bioactivity of the mature toxin.
Fig. 6.
Phylogenetic tree of agatoxins and the remipede neurotoxin. The RAxML likelihood tree of the Agatoxin (RAxML-HPC PHTREADS-SSE3-7.2.6, 10.000 Bootstraps, -f a, PROTGAMMAIWAG, see Materials and Methods). Nonvenomous and nonhematophagous taxa are indicated by asterisks. For descriptions and color code see previous figures. Venom gland transcripts and complete animal tissue transcripts are indicated by the abbreviations VgT and BVgT in parentheses following the sequence name, respectively. Remipede sequences are illustrated by miniaturized remipede pictures, and venom gland pictograms highlight remipede venom gland sequences.
These insights underline the importance of comparing transcripts expressed in the venom glands of species with those expressed in their nonvenom gland tissue (Casewell et al. 2012). To extend and confirm these insights in future work, it will be necessary to profile expression of agatoxin homologs in nonvenom gland tissue in spiders and centipedes, as well as expression of putative homologs in the venom glands of the venomous hymenopterans included in our tree.
Given that the neurotoxins expressed in both spider and centipede venom glands function as ion channel modulators (Billen et al. 2010; Liu et al. 2012), we speculate that the remipede venom gland neurotoxin has a similar activity, causing the immobilization of prey. The adaptive value of a paralyzing neurotoxin in the venom of a blind predator living in nutrient-poor underwater caves would be significant.
Remaining Venom Toxins and Remipede Mode of Feeding
The remaining toxins expressed in the remipede venom glands (fig. 2) include metalloproteases, venom serine carboxypeptidases, serine protease inhibitors (serpin and kunitz), and cysteine-rich secretory proteins related to venom allergen 5 (antigen 5), a major allergen in hymenopteran venom (Moreau 2012). These toxins may contribute a mixture of tissue damaging and ion channel activity modulating activities to the remipede venom. For further discussion of these toxins, we refer the reader to the supplementary material, Supplementary Material online.
A laboratory study of remipede feeding suggested that they may feed predominantly on small particles filtered from the water column (Koenemann et al. 2007) and that remipedes are not obligatory carnivores. We cannot rule out that remipedes in the wild feed on small suspended particles, but the sophisticated functional morphology of their venom delivery system and the expression of a potent mix of tissue damaging enzymes and ion channel activity modulating neurotoxins are unmistakable predatory adaptations. Moreover, laboratory observations (Koenemann et al. 2007) show that remipedes can catch and overpower several types of crustacean prey. This combination of traits together with the presence of a very muscular esophagus (Schram and Lewis 1989; Felgenhauer et al. 1992) support the suggestion, supported by a field observation (Schram and Lewis 1989), that remipedes can feed in an arachnoid manner, sucking the prey’s liquefying tissue out of its cuticle (Schram and Lewis 1989). Remipedes co-occur with a diversity of potential, mostly crustacean, prey, including ostracodes, decapods, amphipods, isopods, mysids, thermosbaenaceans, and copepods (Neiber et al. 2011), and they have been observed carrying ostracodes and consuming caridean shrimp (Schram and Lewis 1989; Carpenter 1999; Koenemann et al. 2007).
Previous Ideas about Remipede Venom Are Likely Erroneous
Our results suggest that the only previously reported evidence on the make-up and effects of remipede venom are probably erroneous (van der Ham and Felgenhauer 2007a, 2007b). van der Ham and Felgenhauer proposed that remipede venom has phenoloxidase activity. They hypothesized that remipede venom glands express a large amount of hemocyanin and an unknown component that could convert the hemocyanin into subunits with phenoloxidase activity. We find no evidence of phenoloxidase expression in either the venom gland or complete animal library, and although the venom gland does express one hemocyanin transcript, it is expressed at an extremely low level (15 reads). In contrast, the complete animal library shows a diversity of hemocyanin transcripts, which is consistent with its role as a respiratory pigment in remipedes (Ertas et al. 2009). We therefore suggest that the phenoloxidase activity observed by van der Ham and Felgenhauer probably represents hemolymph contamination of their venom gland homogenates.
Conclusions
Our results constitute the first conclusive evidence that remipedes are venomous crustaceans. The 3D reconstruction of the venom apparatus of S. tulumensis reveals an effective venom delivery system that is able to separate limb movement and venom injection by the maxillules. Our study also shows the great value of transcriptomic profiling to identify putatively venomous organisms and to generate data on venom composition. The transcriptomic profile of the venom glands of S. tulumensis shows that the injected cocktail of toxins is dominated by enzymes and that it includes a probable paralytic neurotoxin.
This study adds a new major animal group to the roster of known venomous animals and provides the first data on venom composition in a primitively aquatic arthropod lineage. The results indicate that remipede venom is strikingly different from that of other arthropophagous arthropods such as spiders, centipedes, and scorpions. The venoms of these three groups are generally much richer in neurotoxic peptides with much smaller masses than the large enzymes that dominate the remipede venom (Liu et al. 2012; King and Hardy 2013). In terms of the dominance of enzymes, especially proteinases, remipede venom is actually more reminiscent of the venom of viperid snakes (e.g., Casewell et al. 2009; Rokyta et al. 2012).
Our phylogenetic analyses underline the importance of including nonvenom-gland-derived sequences from venomous taxa and sequences from nonvenomous taxa for reconstructing the evolutionary origins and dynamics of venom toxins (Casewell et al. 2012). Without including data derived from a whole animal transcriptomic library, it would have been impossible to infer the full diversity and relationships of nontoxin paralogs expressed in nonvenom gland tissue, or to infer the likely evolutionary origin of the neurotoxin expressed in the remipede venom glands.
This contribution to increasing our knowledge of the toxin composition of independently evolved venoms is crucial for achieving venomics’ ultimate goal of disentangling the phylogenetic, ecological, and other factors that shape the biology and evolution of animal venoms. Additionally, our results are of general importance for future studies of arthropod venom evolution. Remipedes are the earliest branching lineage of venomous pancrustaceans, and as the possible sister group to Hexapoda, they represent the nearest venomous outgroup to the great diversity of venomous insects.
Materials and Methods
Species Collection and Preservation
Specimens of S. tulumensis were collected in several cave diving expeditions to the Yucatan, Mexico. The maxillular glands of 30 specimens were dissected under sterile conditions in RNAlater (supplementary figs. S1 and S5, Supplementary Material online) and used to generate the venom gland tissue-specific transcripts (VgT) on a 454 Titanium platform (Roche). Specimens of different sizes were included in the VgT transcriptome library, which therefore represents different stages and conditions of venom glands. To recover potential nontoxin paralog sequences in nonvenom gland tissue, we analyzed a previously sequenced transcriptome of samples mainly comprising nonvenom gland body tissue (BVgT). BVgT was generated by sequencing a normalized cDNA library on the 454 Titanium platform (Roche), generated from 10 complete specimens that were ground in RNAlater.
Library Reconstruction
Total RNA of dissected venom gland tissue only (VgT) was extracted with the Trizol-GTI-LiCl method and cDNA synthesis, and amplification was conducted with the Mint kit (Evrogen) at LGC Genomics, Berlin, Germany. After cDNA digestion, fragments were size selected on an low melting point (LMP) agarose gel. To prevent exclusion of shorter toxin sequences, the size of selected fragments was lowered to 200 bp. Purified fragments (MinElute Gel extraction kit, Qiagen) were ligated to a pDNR-lib vector (Clontech) using the Fast Ligation Kit (NEB). Half of roughly 2 million plated clones were used for plasmid DNA preparation following standard methods (Qiagen). cDNA inserts were LMP agarose gel purified (MinElute gel extraction kit) and ligated to high-molecular weight DNA using a proprietary Sfi-linker. Library generation was carried out according to the manufacturer’s standard protocols (Roche 454 Life Sciences, Branford, CT), shearing concatenated inserts randomly by nebulization. Sheared fragments were finally added to the fragment ends by ligation. The resulting fragment library was sequenced on 1/4 picotiterplate on the GS FLX using the Roche 454 Titanium chemistry obtaining a total of 260,172 sequence reads.
For the whole animal library, total RNA was extracted from 10 complete specimens, including venom glands (BVgT), with the absolutely RNA kit (Stratagene) and its corresponding cDNA synthesized with the Mint kit (Evrogen) at the Max Planck Institute for Molecular Genetics (MPIMG), Berlin, Germany. A complete 454 FLX Titanium run (Roche) was performed yielding 1,000,000 reads. For further details of sequence processing, see von Reumont et al. (2012).
Sequence Assembly
Before assembly, the VgT sequence reads were screened for the Sfi-linker that was used for concatenation, the linker sequences were clipped out of the reads, and the clipped reads assembled into individual transcripts using the Roche 454 Newbler software at default settings (454 Life Sciences Corporation, Software Release: 2.3), vector sequences were identified and removed using VecScreen. The BVgT reads were assembled as described in von Reumont et al. (2012).
Annotation and Identification of Venom Protein Sequences
To identify venom candidate proteins, we developed a customized bioinformatic pipeline (see supplementary fig. S1, Supplementary Material online). First, the transcriptome sequences were translated in amino acids for all six reading frames. This is needed to cover possible protein variants on different strands or in different reading frames. The six-frame translated protein sequences were formatted into a local Blast database to search for different known proteins. Venom proteins are known to be usually represented by secreted proteins, so we blasted all secreted proteins from UniProt (location_sl_0243) against our translated cDNA databases. The results of the Blast search were then parsed into a hit-sequence file, including all sequences with Blast hits for secretory proteins, which was used in all further analyses to identify putative venom proteins. A functional annotation and identification of active sites and conserved motives was conducted by feeding the hit-sequence file into Blast2GO (Conesa and Götz 2008). This software allowed a final, automated InterProscan for active sites and motifs (Hunter et al. 2012), which we used in addition to standard Blast procedures to predict putative venom proteins. General sequence annotation was performed with Blasthits (nr), InterProscan and Gene Ontology term annotation from Blast2GO (see supplementary tables S1 and S2, Supplementary Material online).
Phylogenetic Reconstruction of Venom Protein Trees
Sequences were aligned using Mafft E-INS-I27 relying, where available, on Uniprot reviewed manually curated venom protein structure constraints (Jungo et al. 2012). Generally, sequences in which stop codons interrupted the open reading frames (potentially representing sequencing artifacts or pseudogenes) were removed from the final alignments. Unless noted otherwise, most of these alignments excluded the prepro regions coding for the signal peptide and propeptides that are posttranslationally removed from the mature (domain region) protein. The prepro regions were mostly too diverse for unambiguous alignment. After chosing the best fitting model with ProtTest 328, a maximum likelihood analysis was conducted with RAxML-HPC PTHREADS-SSE3-7.2.6 (Stamatakis and Alachiotis 2010) for each of the protein alignments using the best fitting protein model to reconstruct phylogenetic trees (see figure legends of reconstructed trees for the chosen model for each protein). Bootstrap support was estimated from 10,000 pseudoreplicates. Unless mentioned otherwise in the figure legends, all trees were rooted with a nonvenom transcript variant from a nonvenomous vertebrate.
Additional sequences from venomous and nonvenomous taxa were predominantly obtained from UniProt to maximize the number of annotated sequences in our analyses. Where possible nonvenomous arthropod species from the four major groups were included.
Three-Dimensional Reconstruction of SR-µCT Data
Two samples preserved in Bouin were transferred to 70% ethanol and subsequently critical point dried (Model E4850, BioRad, Hercules, CA) to reduce fluid induced artifacts during the scanning process. Subsequently samples were superglued on specimen holders for SR-µCT (Betz et al. 2007). SRµ-CT has a high penetrating power and allows visualizing large specimens without the need for sectioning. Scanning was conducted at the German electron synchrotron accelerator (DESY, Hamburg, Germany; the tomography station BW2/DORIS III operated by Helmholtz-Zentrum Geesthacht HZG, Geesthacht, Germany), which is optimized for performing high-density resolution microtomography (Beckmann et al. 2008). The facility provides floating point data as well as 16 bit TIFF image files and volume data files (.vgi-format) ready for analysis. The provided volume data (.vgi-files) was analyzed with the free myVGL 2.0 64 bit viewer (Volume Graphics, Heidelberg, Germany). Segmentation and rendering of single structures was accomplished with Reconstruct (Fiala 2005) and Blender (http://www.blender.org, last accessed November 4, 2013). Final figures were edited with GIMP, Inkscape, and Scribus (all GPL).
Supplementary Material
Supplementary material figures S1–S9 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
B.M.v.R. and R.A.J. designed the study. B.M.v.R. collected the specimens. B.M.v.R., C.B., and S.R. performed the bioinformatic analyses. B.M.v.R., R.A.J., and C.B. led data interpretation. A.B. and B.M.v.R. performed the synchrotron scan of the specimens. A.B. reconstructed the specimen in 3D. All authors wrote and approved the manuscript. Berthold Fartmann (LGC Genomics, Berlin, Germany) was tremendously helpful in generating the venom gland transcripts. Lars Hering (University of Leipzig) helped to set up our bioinformatic pipelines. Phylogenetic analyses were performed with the assistance of SBCS Informatics (http://informatics.sbcs.qmul.ac.uk), the EPRSC funded MidPlus cluster at Queen Mary University in London, United Kingdom, and the cluster system at the Center of Molecular Biodiversity (CMB), ZFMK Bonn, Germany. We thank Fred Schram and Stefan Koenemann for discussing remipede morphology. Bryan Fry was helpful in discussing venom-related questions. Lahcen Campell provided useful comments on the manuscript and scripts. Julie Neisch deserves special thanks as safe cave diving buddy of BMvR while collecting specimens. Tom M. Iliffe (TAMUG) and especially Brett Gonzalez helped to collect specimens. For funding B.M.v.R. thanks DFG (grants RE 3454/1-1, WA 530/34), Raiffeisenbank Bonn Rhein Sieg, Germany, and for sponsoring and support he thanks Poseidon Deutschland, and Thorsten Waelde and Bruno Spagnuolo of the technical cave diving training facility Protec Sardinia, Italy. C.B. received funding from the DFG (BL787/7-1). We are further grateful to Felix Beckmann (HZG, Geesthacht, Germany) for guaranteeing high-quality SR-µCT scans. Karen Meusemann and Hans Meusemann were great helps as scanning team members. SR-µCT scans at DESY Hamburg, Germany, were financed by HASYLAB grant I-20080169 to A.B. R.A.J. gratefully acknowledges support from NERC (grant NE/I001530/1) and BBSRC (grant BB/K003488/1).
References
- Almeida DD, Scortecci KC, Kobashi LS, Agnez-Lima LF, Medeiros SRB, Silva-Junior AA, Junqueira-De-Azevedo I, Fernandes-Pedrosa M. Profiling the resting venom gland of the scorpion Tityus stigmurus through a transcriptomic survey. BMC Genomics. 2012;13:362. doi: 10.1186/1471-2164-13-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminetzach Y, Srouji J, Kong C, Hoekstra H. Convergent evolution of novel protein function in shrew and lizard venom. Curr Biol. 2009;19:1925–1931. doi: 10.1016/j.cub.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian PG, Beckmann A, Warnken U, Schnolzer M, Schuler A, Bornberg-Bauer E, Holstein TW, Ozbek S. Proteome of Hydra nematocyst. J Biol Chem. 2012;287:9672–9681. doi: 10.1074/jbc.M111.328203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann F, Herzen J, Haibel A, Müller B, Schreyer A. High density resolution in synchrotron-radiation-based attenuation-contrast microtomography. Proc SPIE. 2008;7078:70781D. [Google Scholar]
- Betz O, Wegst U, Weide D, Heethoff M, Helfen L, Lee WK, Cloetens P. Imaging applications of synchrotron X-ray phase-contrast microtomography in biological morphology and biomaterials science. I. General aspects of the technique and its advantages in the analysis of millimetre-sized arthropod structure. J Microsci. 2007;227:51–71. doi: 10.1111/j.1365-2818.2007.01785.x. [DOI] [PubMed] [Google Scholar]
- Billen B, Vassilevski A, Nikolsky A, Debaveye S, Tytgat J, Grishin E. Unique bell-shaped voltage-dependent modulation of Na+ channel gating by novel insect-selective toxins from the spider Agelena orientalis. J Biol Chem. 2010;285:18545–18554. doi: 10.1074/jbc.M110.125211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust A, Sunagar K, Undheim EAB, et al. (15 co-authors) Differential evolution and neofunctionalization of snake venom metalloprotease domains. Mol Cell Proteomics. 2013;12:651–663. doi: 10.1074/mcp.M112.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter J. Behavior and ecology of Speleonectes epilimnius (Remipedia, Speleonectidae) from surface water of an anchialine cave on San Salvador Island, Bahamas. Crustaceana. 1999;72:979–991. [Google Scholar]
- Casewell NR, Harrison RA, Wüster W, Wagstaff SC. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics. 2009;10:564. doi: 10.1186/1471-2164-10-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Huttley GA, Wüster W. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun. 2012;3:1066–1010. doi: 10.1038/ncomms2065. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wagstaff SC, Harrison RA, Renjifo C, Wuster W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Mol Biol Evol. 2011;28:2637–2649. doi: 10.1093/molbev/msr091. [DOI] [PubMed] [Google Scholar]
- Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends Ecol Evol. 2013;28:219–229. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhao L, Jiang L, Meng E, Zhang Y, Xiong X, Linag S. Transcriptome analysis revealed novel possible venom components and cellular processes of the tarantula Chilobrachys jingzhao venom gland. Toxicon. 2008;52:794–806. doi: 10.1016/j.toxicon.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Pinto JRA, Fox EGP, Saidemberg DM, Santos LD, da Silva Menegasso AR, Costa-Manso E, Machado EA, Bueno OC, Palma MS. Proteomic view of the venom from the fire ant Solenopsis invicta Buren. J Proteome Res. 2012;11:4643–4653. doi: 10.1021/pr300451g. [DOI] [PubMed] [Google Scholar]
- Ertas B, von Reumont BM, Wägele JW, Misof B, Burmester T. Hemocyanin suggests a close relationship of Remipedia and Hexapoda. Mol Biol Evol. 2009;26:2711–2718. doi: 10.1093/molbev/msp186. [DOI] [PubMed] [Google Scholar]
- Fanenbruck M, Harzsch S, Wägele JW. The brain of the Remipedia (Crustacea) and an alternative hypothesis on their phylogenetic relationships. Proc Natl Acad Sci U S A. 2004;101:3868–3873. doi: 10.1073/pnas.0306212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgenhauer BE, Abele LG, Felder DL. Remipedia. In: Harrison FW, Humes AG, editors. Microscopic anatomy of invertebrates. Vol. 9. Crustacea. New York: Wiley-Liss; 1992. pp. 225–247. [Google Scholar]
- Fernandes-Pedrosa M, Junqueira-De-Azevedo I, Gonçalves-De-Andrade R, Kobashi L, Almeida D, Ho P, Tambourgi D. Transcriptome analysis of Loxosceles laeta (Araneae, Sicariidae) spider venomous gland using expressed sequence tags. BMC Genomics. 2008;9:279. doi: 10.1186/1471-2164-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC. Reconstruct: a free editor for serial section microscopy. J Microsci. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Fry B, Roelants K, Champagne DE, et al. (12 co-authors) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu Rev Genomics Hum Genet. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- Fry BG, Winter K, Norman JA, et al. (22 co-authors) Functional and structural diversification of the Anguimorpha lizard venom system. Mol Cell Proteomics. 2010;9:2369–2390. doi: 10.1074/mcp.M110.001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet G, Edgecombe GD. Reevaluating the arthropod tree of life. Annu Rev Entomol. 2012;57:167–186. doi: 10.1146/annurev-ento-120710-100659. [DOI] [PubMed] [Google Scholar]
- Hölldobler B, Wilson EO. The superorganism. The beauty, elegance, and strangeness of insect societies. New York: W. W. Norton & Company; 2009. [Google Scholar]
- Hunter S, Jones P, Mitchell A, et al. (50 co-authors) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungo F, Bougueleret L, Xenarios I, Poux S. The UniProtKB/Swiss-Prot Tox-Prot program: a central hub of integrated venom protein data. Toxicon. 2012;60:551–557. doi: 10.1016/j.toxicon.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GF. Venoms as a platform for human drugs: translating toxins into therapeutics. Expert Opin Biol Ther. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- King GF, Hardy MC. Spider-venom peptides: structure, pharmacology, and potential for control of insect pests. Annu Rev Entomol. 2013;58:475–496. doi: 10.1146/annurev-ento-120811-153650. [DOI] [PubMed] [Google Scholar]
- Koenemann S, Schram FR, Iliffe TM, Hinderstein LM, Bloechl A. Behavior of Remipedia in the laboratory, with supporting field observations. J Crust Biol. 2007;27:534–542. [Google Scholar]
- Lin H-H, Tseng L-H. DBCP: a web server for disulfide bonding connectivity pattern prediction without the prior knowledge of the bonding state of cysteines. Nucleic Acids Res. 2010;38:W503–W507. doi: 10.1093/nar/gkq514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z-C, Zhang R, Zhao F, et al. (12 co-authors) Venomic and transcriptomic analysis of centipede Scolopendra subspinipes dehaani. J Proteome Res. 2012;11:6197–6212. doi: 10.1021/pr300881d. [DOI] [PubMed] [Google Scholar]
- Martin JW, Davis GE. An updated classification of the Recent Crustacea. Nat Hist Mus Los Angel Cty Sci Ser. 2001;39:1–124. [Google Scholar]
- Moreau SJ. “It stings a bit but it cleans well”: venoms of Hymenoptera and their antimicrobial potential. J Insect Physiol. 2012;59:186–204. doi: 10.1016/j.jinsphys.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Morgenstern D, Rohde BH, King GF, Tal T, Sher D, Zlotkin E. The tale of a resting gland: transcriptome of a replete venom gland from the scorpion Hottentotta judaicus. Toxicon. 2011;57:695–703. doi: 10.1016/j.toxicon.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Neiber MT, Hartke TR, Stemme T, Bergmann A, Rust J, Iliffe TM, Koenemann S. Global biodiversity and phylogenetic evaluation of Remipedia (Crustacea) PLoS One. 2011;6:e19627. doi: 10.1371/journal.pone.0019627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 2010;463:1079–1083. doi: 10.1038/nature08742. [DOI] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K. The venom-gland transcriptome of the eastern diamondback rattlesnake (Crotalus adamanteus) BMC Genomics. 2012;13:312. doi: 10.1186/1471-2164-13-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruder T, Sunagar K, Undheim EAB, Ali SA, Wai T-C, Low DHW, Jackson TNW, King GF, Antunes A, Fry BG. Molecular phylogeny and evolution of the proteins encoded by coleoid (cuttlefish, octopus, and squid) posterior venom glands. J Mol Evol. 2013;76:192–204. doi: 10.1007/s00239-013-9552-5. [DOI] [PubMed] [Google Scholar]
- Schram FR, Hof CHJ. Fossils and interrelationships of major crustacean groups. In: Edgecombe GD, editor. Arthropod fossils and phylogeny. New York: Columbia University Press; 1998. pp. 233–302. [Google Scholar]
- Schram FR, Lewis CA. Functional morphology of feeding in the Nectiopoda. In: Felgenhauer BE, Watling L, Thistle AB, editors. Crustacean issues 6. Functional morphology of feeding and grooming in Crustacea. Rotterdam (The Netherlands): Balkema; 1989. pp. 115–122. [Google Scholar]
- Stamatakis A, Alachiotis N. Time and memory efficient likelihood-based tree searches on phylogenomic alignments with missing data. Bioinformatics. 2010;26:i132–i139. doi: 10.1093/bioinformatics/btq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagar K, Johnson WE, O'brien SJ, Vasconcelos V, Antunes A. Evolution of CRISPs associated with toxicoferan-reptilian venom and mammalian reproduction. Mol Biol Evol. 2012;29:1807–1822. doi: 10.1093/molbev/mss058. [DOI] [PubMed] [Google Scholar]
- van der Ham JL, Felgenhauer BE. The functional morphology of the putative injecting apparatus of Speleonectes tanumekes (Remipedia) J Crust Biol. 2007a;27:1–9. [Google Scholar]
- van der Ham JL, Felgenhauer BE. On possible venomous effects of Speleonectes sp. (Remipedia) Crustaceana. 2007b;80:755–765. [Google Scholar]
- Vincent B, Kaeslin M, Roth T, et al. (11 co-authors) The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genomics. 2010;11:693. doi: 10.1186/1471-2164-11-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reumont BM, Burmester T. In: Encyclopedia of Life Sciences (ELS) Chichester: John WIley & Sons, ltd; 2010. Remipedia and the evolution of hexapods. [Google Scholar]
- von Reumont BM, Jenner RA, Wills MA, et al. (13 co-authors) Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the sister group of Hexapoda. Mol Biol Evol. 2012;29:1031–1045. doi: 10.1093/molbev/msr270. [DOI] [PubMed] [Google Scholar]
- Whittington CM, Papenfuss AT, Locke DP, et al. (12 co-authors) Novel venom gene discovery in the platypus. Genome Biol. 2010;11:R95. doi: 10.1186/gb-2010-11-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager J. Remipedia, a new class of Crustacea from a marine cave in the Bakamas. J Crust Biol. 1981;1:328–333. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.