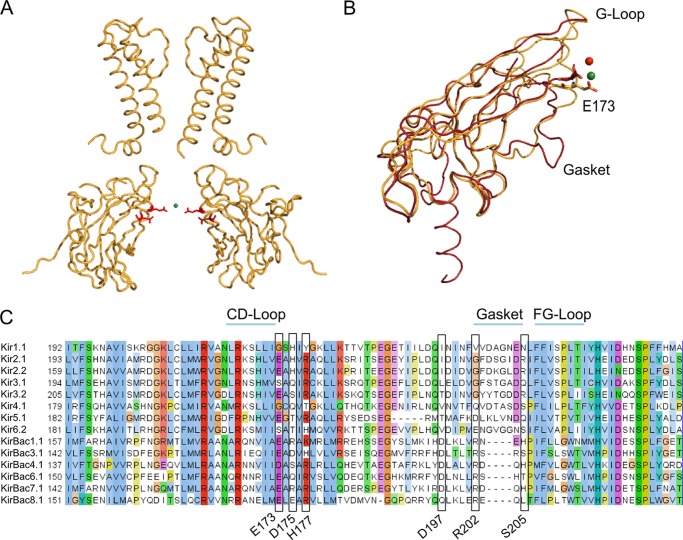
FIGURE 4.
Ion coordination sites. A, the positions of Glu173 and Asp175 (red sticks) in KirBac3.1. The potassium ion within the cavity (green sphere) appears to be coordinated by these residues. For clarity, only two subunits are shown. B, structural alignment of the cytoplasmic domains of KirBac3.1 S129R/S205L (gold) and Kir3.1 S225E (PDB ID 3K6N; red). The Na+ ion identified in the Kir3.1 S225E structure is shown as a red sphere, whereas the K+ ion in KirBac3.1 S129R/S205L is shown as a green sphere. Mutation S225E in Kir3.1 mimics the Glu224/5 residue found in strongly rectifying Kir2.1 and Kir2.2. This negatively charged residue, along with the gasket loop, provides the cytoplasmic pore with cation coordination sites. They play an important role in facilitating polyamine block and in maintaining normal channel conductance. The gasket loop is not found in prokaryotic Kir channels. C, Glu173 in KirBac3.1 is highly conserved in both eukaryotic and prokaryotic channels. Other residues relevant to this study are also highlighted.