FIGURE 2.
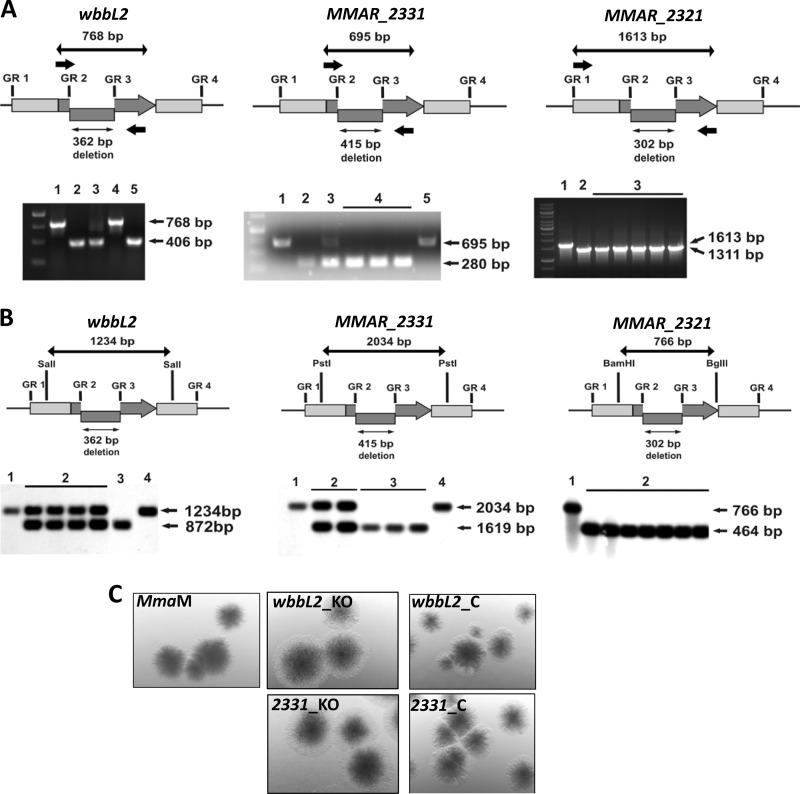
Gene inactivation of wbbL2, MMAR_2331, and MMAR_2321 in M. marinum. A, PCR-based genotyping of the mutants. Top, schematic showing the genomic orientation of the wbbL2, MMAR_2331, and MMAR_2321 genes (thick gray arrows). The internal deletion introduced into a gene using a gene replacement vector is marked with a shifted rectangle. GR1-GR2/GR3-GR4 denote the gene-flanking fragments that were amplified and cloned together in the pGR2349, pGR2331, or pGR2321 suicide vector to obtain the mutated ΔwbbL2, ΔMMAR_2331, and ΔMMAR_2321 allele for gene replacement. The small thick black arrows indicate the binding sites for the primers that were used for PCR-based genotyping. The thick black arrows indicate the length of the product amplified from wild-type wbbL2 (768 bp), MMAR_2331 (695 bp), and MMAR_2321 (1613 bp). The thin arrows represent the length of the deleted fragment (362 bp, yielding a 406-bp ΔwbbL2 PCR product; 415 bp, yielding a 280-bp ΔMMAR_2331 PCR product; 302 bp, yielding a 1311-bp PCR product). Bottom, the genotypes of selected M. marinum wbbL2, MMAR_2331, and MMAR_2321 mutants were confirmed by PCR. For wbbL2, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2349; lane 3, SCO strain carrying both wild-type and mutated genes; lane 4, the WT-DCO strain; lane 5, DCO strain ΔwbbL2. For MMAR_2331, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2331; lane 3, SCO strain; lane 4, DCO strain ΔMMAR_2331; lane 5, WT-DCO strain. For MMAR_2321, lanes are as follows: lane 1, control PCR conducted on wild-type M. marinum DNA; lane 2, control PCR conducted on pGR2321; lane 3, DCO knock-out strain ΔMMAR_2321. B, Southern blot analysis of the mutants. In the schematics (upper panels), the long black arrow represents the restriction-digested DNA fragment (1234 bp for wbbL2, 2034 bp for MMAR_2331, and 766 bp for MMAR_2321), whereas the short black arrow indicates the size of the internal deletion in the mutated gene (362 bp for ΔwbbL2, 415 bp for ΔMMAR_2331, and 302 bp for ΔMMAR_2321). The wbbL2, MMAR_2331, and MMAR_2321 genes are represented by the gray arrows, and the internal deletion is represented by the shifted rectangles. Shown is a Southern blot (lower panels) confirming the deletion in mutated M. marinum strains. Lanes are as follows: for ΔwbbL2 and ΔMMAR_2331: lane 1, wild-type M. marinum; lane 2, SCO strains; lane 3, DCO knock-out ΔwbbL2 or ΔMMAR_2331 mutant; lane 4, wild-type DCO; for ΔMMAR_2321: lane 1, wild-type M. marinum; lane 2, DCO knock-out ΔMMAR_2321. C, effect of deletion of wbbL2 or MMAR_2331 on colony morphology. Shown are colonies of M. marinum strain M (wild type), wbbL2, or MMAR_2331 deletion mutants and their corresponding complemented strains (wbbL2_C and 2331_C) on 7H10 agar plates. Compared with wild-type M. marinum strain M, both mutants presented a rough and dried texture, a phenotype that was restored following complementation.