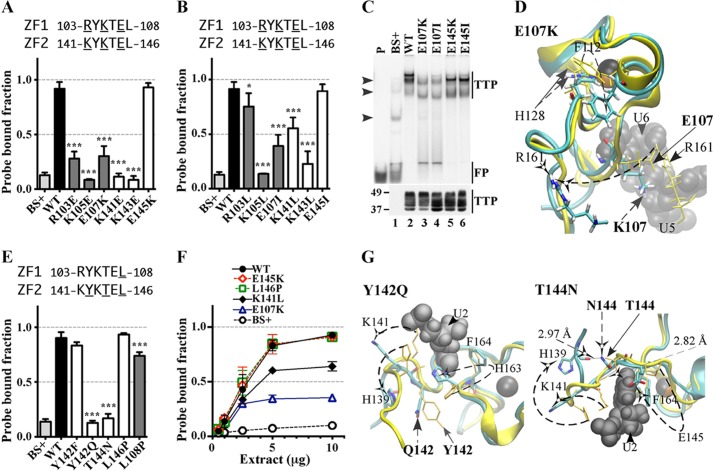
FIGURE 5.
Effects of substitution mutants in the lead-in sequences to the zinc fingers on RNA binding. The binding of protein to RNA is expressed as the probe-bound fraction. Above the graphs are the amino acid sequences of the lead-in amino acids. The underlined residues are those that were mutated and whose effects on binding are shown in the bar graphs. The results from mutants of finger 1 are shown as gray columns, and those of finger 2 are shown as white columns. A and B, effects of mutant and WT TTP proteins, expressed as the probe-bound fraction. A, effects of charge reversal mutants; B, effects of neutral mutants. The results (mean ± S.D. (error bars)) are from three similar gel shift assays. C, the top panel shows representative gel shift assays using extracts from HEK293 cells transfected with vector alone (BS+), WT human TTP, or various mutant TTP constructs. Cytosolic extracts were incubated with 0.2 nm probe TARE5 as described under “Experimental Procedures” before loading on a non-denaturing acrylamide gel. A sample of probe alone in buffer was also loaded (lane P). Migration of the TTP-ARE complex (TTP) and the free probe (FP) are indicated on the right by vertical bars, and the three major endogenous protein-ARE complexes are labeled with arrowheads on the left. The bottom panel shows the relative amount of WT TTP protein and its mutants used in the binding reaction, as determined by Western blotting. D, superposition of WT TTP finger 1 and mutant E107K finger 1 structural ensembles (RMSD is 6.24 Å; RMSD values were calculated from superposition of the TZF domains of each mutant TTP with RNA-bound WT TTP). Wild-type TTP peptide backbone ribbon and side chains are in yellow, mutant E107K peptide backbone ribbon is in cyan, and side chains are in colors. RNA nucleosides U5 and U6 are shown as gray spheres. Zinc atoms are shown as black and copper spheres. E, comparisons in RNA binding are shown between mutant and WT TTP proteins; the results (mean ± S.D.) are from three similar gel shift assays. F, binding assays with 0.2 nm TARE5 probe in serially diluted extract with 1–10 μg of total protein. The probe-bound fractions (mean ± S.D.) are from three similar gel shift assays. G, superposition of WT TTP finger 2 and the Y142Q (RMSD value is 7.41 Å) or T144N (RMSD value is 10.66 Å) mutant model peptides. Color schemes are the same as in C, and RNA nucleoside U2 is shown. Zinc atoms are shown as silver and copper spheres.