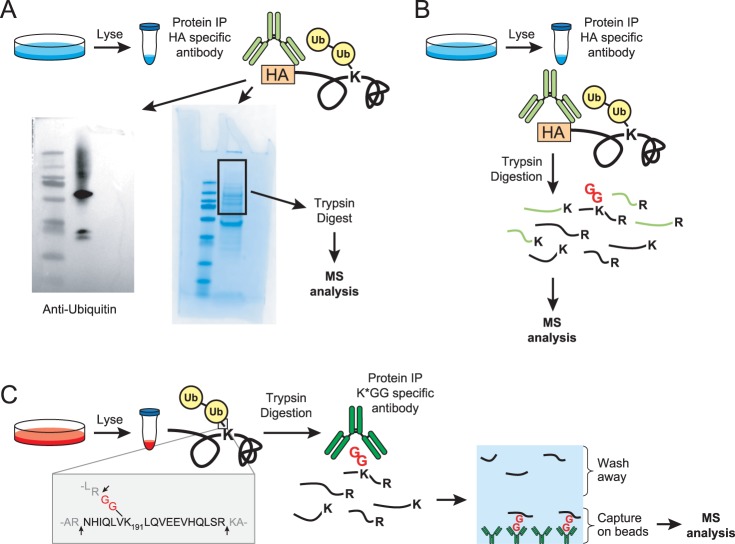
Fig. 1.
Methods to map ubiquitination sites. A, Schematic of the gel-based method for identifying ubiquitination sites on proteins of interest (POI). Proteins are immunoprecipitated using an immunoaffinity tag or with an antibody specific for a POI followed by SDS-PAGE analysis. Anti-ubiquitin Western blots are used to confirm the location of ubiquitinated POI and Coomassie stained gel bands are cut accordingly. In-gel tryptic digestions are performed, peptides are extracted, and then analyzed by LC-MS/MS. This example shows a representative Western blot and Coomassie from the HA-tagged TCRα studies. B, In solution digestion method on a POI. This is the same as (A) except trypsin digestions are performed immediately post-IP (in the presence of the IP antibody) and no SDS-PAGE analysis is performed. C, Schematic of the K-GG peptide immunoaffinity enrichment method for mapping ubiquitination sites on a POI. Whole cell lysates are digested with trypsin and the resultant peptides are immunoprecipitated with an antibody against the K-GG remnant left on ubiquitinated lysine residues. Peptides are eluted and analyzed by LC-MS/MS.