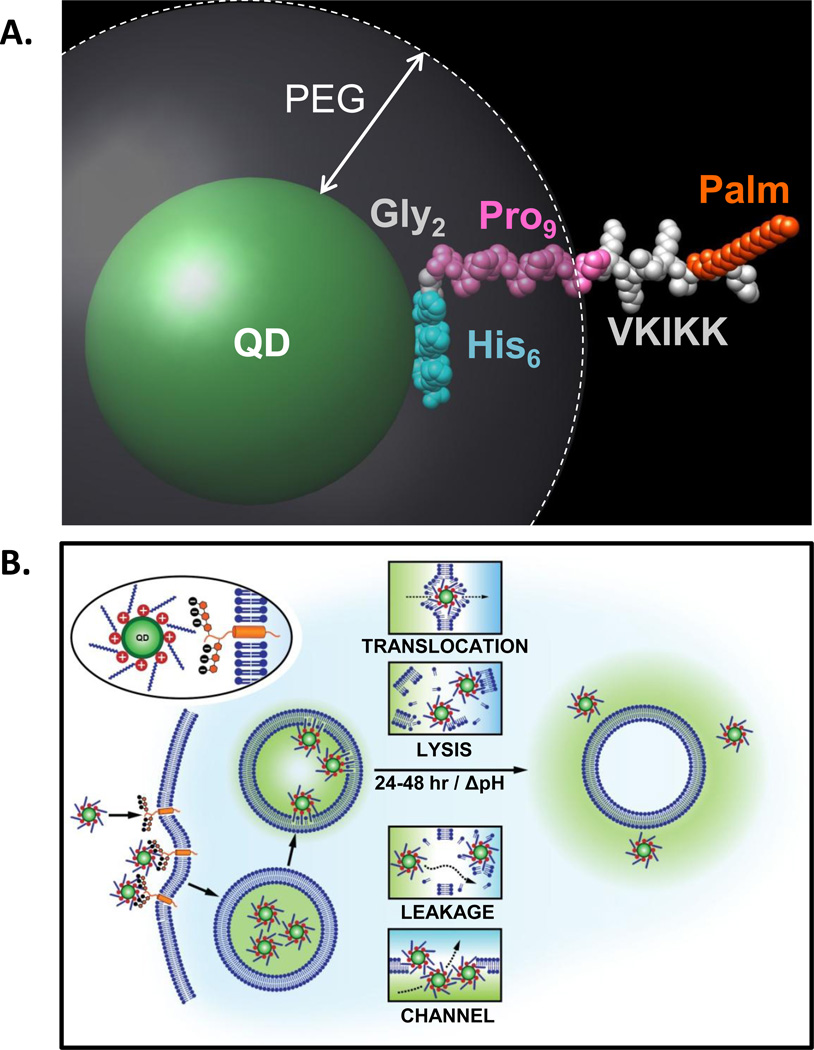
Figure 6. Model of JB577-mediated cytosolic escape of QDs.
(A) Simulation of JB577 structure on 550 nm emitting QDs. The 550 nm QD core/shell diameter (~56Å) and the extension of the PEG ligand on the QD surface (~30Å) were derived in refs.26,43 The His6 sequence (light blue) is assumed to be in contact with the QD surface and does not contribute to lateral extension. This is followed by the Gly2 flexible linker (grey) and the Pro9 motif (pink) forms a rigid type II helix. The QD-assembled conformation and extension of the His6Gly2Pro9 portion has been previously confirmed with FRET.44, 45, 49 The VKIKK sequence is then depicted in grey outside the PEG layer along with the palmitoyl (orange) suggesting that both are available for interactions with the cell membrane. See also SI. (B) Schematic depicting putative steps in the endocytic uptake of QD-JB577 conjugates. Initial interactions are mediated by the positively charged Lys residues and negatively-charged heparan proteoglycan sulfates. Following endosomal formation, the palmitoyls surrounding the QDs penetrate the vesicular membrane. During the next 24–48 h, the QD-peptido-conjugates escape the endosomes in a relatively non-toxic manner and some potential mechanisms are suggested