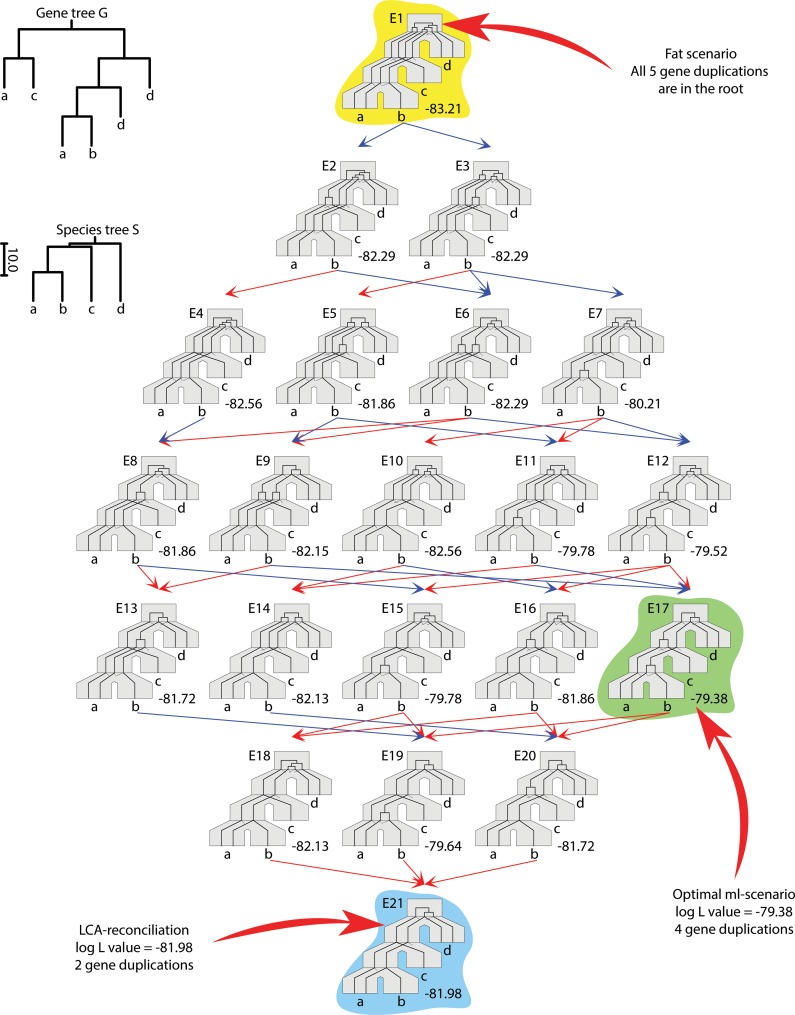
FIG. 1.
A gene tree G = ((a,c), (((a,b),d),d)) and a species tree S = (((a : 9, b : 9) : 8,c : 17) : 1,d : 18) : 2 with evolutionary scenarios (embeddings) in the duplication-loss model are depicted as a reduction diagram (for more details, see Górecki and Tiuryn, 2006). The log L values are computed for λ = 1.3. The most likely evolutionary scenario is E17 (log L = −78.38). DrML can compute this value as well as the optimal DLS tree(s). Additionally, DrML computes the number of gene duplications and log L values for the lca scenario (the minimal scenario in the duplication-loss model; see the bottom embedding E21) and for the fat scenario (the scenario having all possible gene duplications in the root of the species tree; see the top embedding E1).