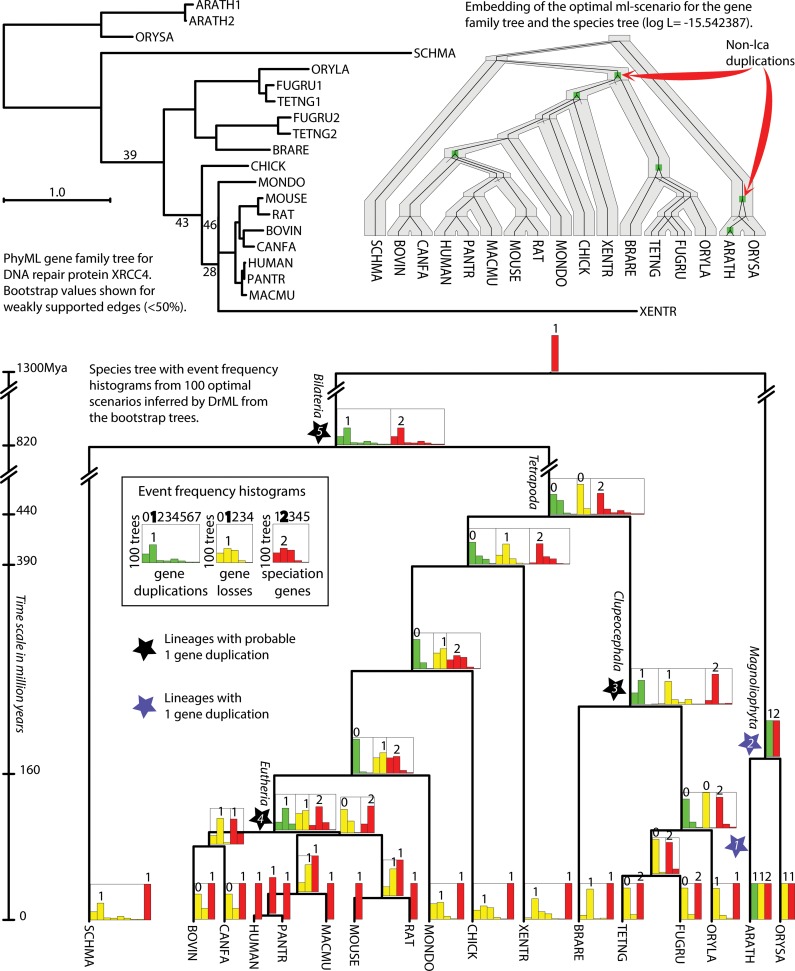
FIG. 3.
Gene duplication analysis by gene tree bootstrapping and using DrML. (Top left) PhyML gene tree for DNA repair proteins XRCC4 rooted by Urec (Górecki and Tiuryn, 2007). Bootstrap values for 100 samples are shown for the edges with the support less than 50%. (Top right) Embedding of the ML scenario inferred by DrML into the species tree (shown below). This optimal scenario has two more duplications than the lca scenario. These non-lca duplications are indicated by red arrows. The log L of the optimal scenario equals −15.542387 (−18.649822 for the lca scenario). (Bottom) The species tree with branch lengths proportional to time with event frequency histograms computed for the set of 100 optimal DrML scenarios inferred for the 100 bootstrap gene trees. For each edge of the species tree, we present three histograms showing the frequencies of given type of events. Each histogram has a number shown above the highest bar. This number denotes the most frequent number of events associated with a given edge among analyzed set of scenarios. Stars denote the edges where gene duplication events are the most likely.