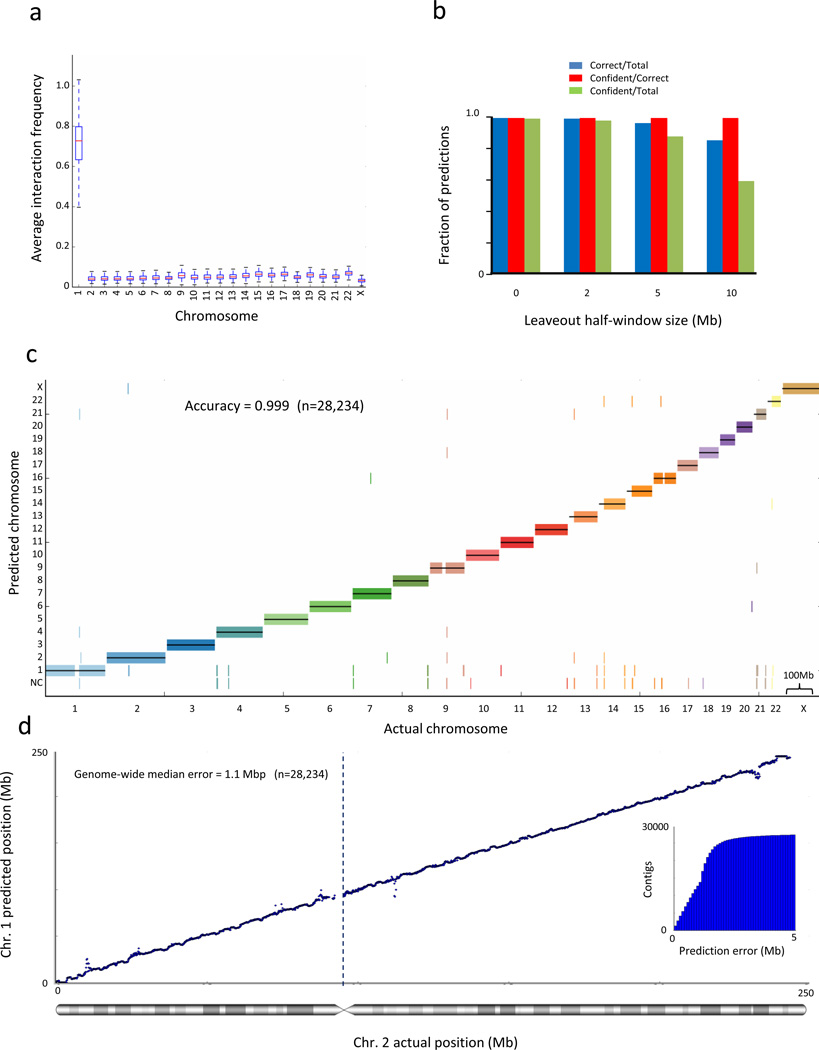
Figure 1.
Interaction frequency accurately predicts chromosome and locus for scaffold augmentation. (a) Average interaction frequency strongly separates interchromosomal from intrachromosomal interactions. For each 100kb contig in chromosome 1, we calculate its average interaction frequency with each chromosome. We exclude interaction data from the contig’s 1 Mb regions on each side, where the strongest interaction frequencies are typically found. The box plot shows the distribution of average interaction frequencies of all contigs over all chromosomes and demonstrates that the distribution of interchromosomal interaction frequencies is separated from intrachromsomal interaction frequencies. Whiskers represent minimal and maximal points within 1.5 of the interquartile range. (b) Naïve Bayes predictive performance at various gap sizes. We trained a Naïve Bayes classifier and predicted the chromosome of each contig, leaving out a 1/2/5/10 Mb flanking region on each side of the contig. The accuracy of all cross-validated predictions and of the confident predictions is shown by the left y-axis and the blue and red lines, respectively. The fraction of total predictions that are confident is shown by the right y-axis and the black line. (c) Genome-wide view of Naïve Bayes predictive performance. The prediction for each contig is marked by a short vertical line, colored according to its true chromosome. Predictions showed were performed leaving out a 1 Mb flanking region on each side of the contig. Predictions that did not pass the confidence threshold are marked as “NC”. (d) Interaction frequencies accurately predict chromosomal locus. For every contig, we exclude interaction data from the contig’s 1Mb flanking regions on each side and then predict its location in cross-validation. The inset shows the cumulative distribution of the absolute prediction error. All statistics are genome-wide.