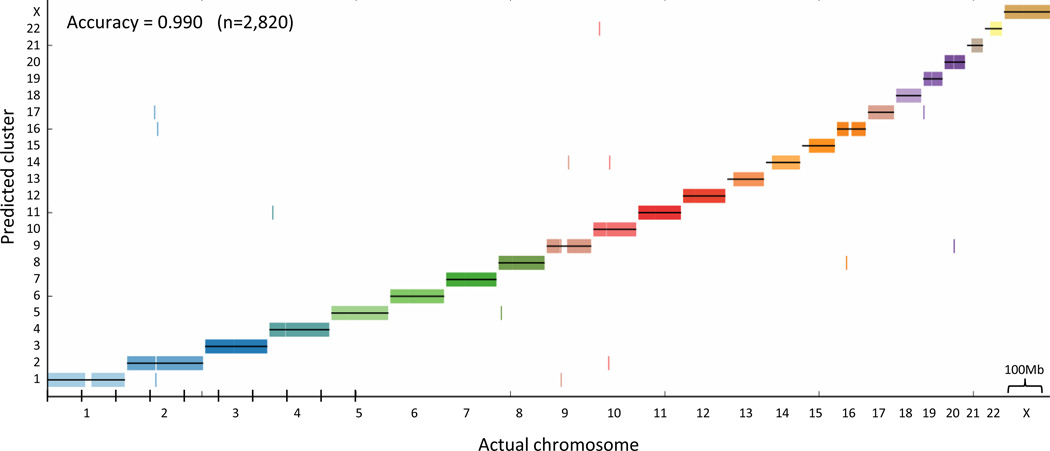
Figure 3.
De novo karyotyping (chromosome assignment). We retained every tenth 100 kb contig in the genome, leaving 0.9 Mb gaps between contigs. We then transformed the interaction frequencies into approximate distances and applied standard average linkage hierarchical clustering to the approximate distance matrix, without using any prior knowledge regarding the positions of the contigs. The cluster assignment for each contig is marked by a short vertical line, colored according to its true chromosome.