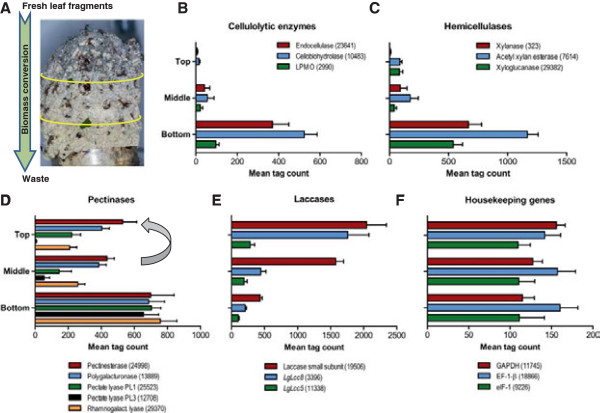
Figure 1.
Fungus garden expression profiles of selected biomass-conversion and housekeeping genes. Genes were identified and their expression profiles obtained by DeepSAGE analysis on the three sections of the ant fungus garden (mean number of mono-tags + SE). The genes are labeled by their inferred gene function with their mono-tag id in parenthesis. (A) A fungus garden of a laboratory colony of the leaf-cutting ant Acromyrmex echinatior. The inverted plastic beaker normally covering the garden has been removed before taking the photo. The subdivision of the garden into three sections (top, middle, bottom) is indicated by the yellow concentric rings. Samples for RNA extraction were taken from the center of each section (Photo courtesy of David Nash, University of Copenhagen). (B) Expression profiles of three genes encoding cellulolytic enzymes attacking the crystalline cellulose microfibrils. LPMOs (lytic polysaccharide monooxygenases) of family AA9 (formerly family GH61) attack the microfibrils using reactive oxygen species. (C) Expression profiles of three genes encoding hemicellulases each containing a CBM1. (D) Expression profiles of five genes encoding pectinolytic enzymes. The pectinesterase [GenBank:HQ174766] and the polygalacturonase [GenBank:HQ174767] were previously identified as two of the major fecal fluid pectinases that the ants transfer from the gongylidium-rich middle section to the top section of the garden (curved arrow) [7]. The pectate and rhamnogalacturonan lyases were not among the fecal fluid enzymes and are thus inferred to be active in the section of the garden where they are expressed. (E) Expression profiles of three genes encoding laccases. The laccases include LgLcc8 [GenBank:JQ307230] and LgLcc5 [GenBank:JQ307227] [8]. The small subunit laccase has not previously been described. (F) Expression profiles of three housekeeping genes that were not significantly differentially expressed among the garden sections (see Results). GAPDH, glyceraldehyde 3-phosphate dehydrogenase; EF-1-β, elongation factor 1-β; eIF-1, eukaryotic translation initiation factor 1.