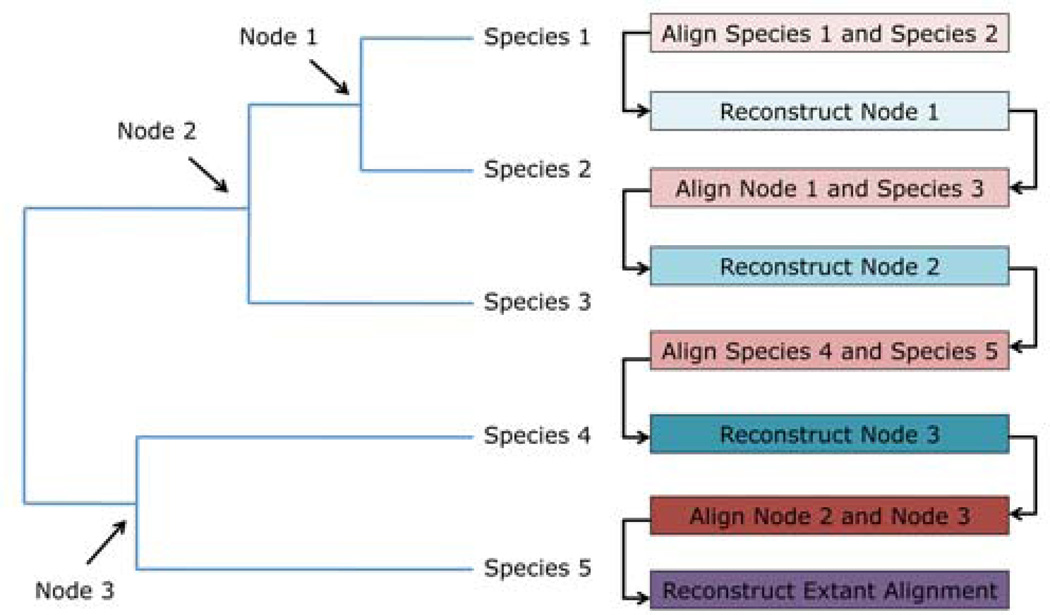
Figure 1. Using ancestral sequence reconstruction to guide alignment.
We aligned the amino acid sequences of the most closely related species to one another, then used PAML (codeml) to reconstruct the ancestral nucleotide sequence for each node (Methods). We continued this process until Nodes 2 and 3 could be aligned to one another. Finally, we remapped the extant sequences onto this alignment.