Figure 1. Girk channel structure.
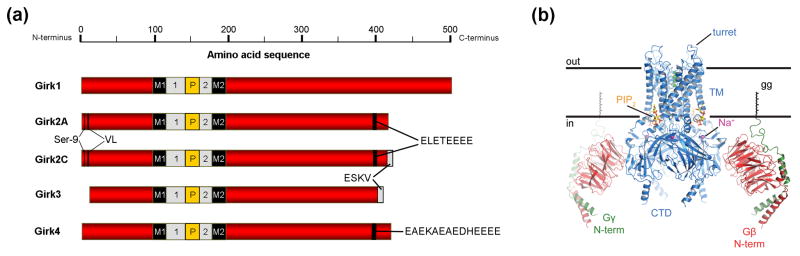
(a) Linear depiction of Girk channel subunits, including two prevalent Girk2 splice variants expressed in the mouse nervous system. The four Girk subunits exhibit a high degree of homology in the membrane-spanning (M1, M2), extracellular (1, 2), and pore (P) domains, with most inter-subunit differences observed in the distal N- and C-terminal domains. The Girk2 splice variants and Girk4 contain ER export signals (ELETEEEE and EAEKAEAEDHEEEE, respectively) not found in the other subunits [7]. Girk2 also exhibits an N-terminal internalization motif (VL), whose influence on channel trafficking is precluded by phosphorylation of Girk2(Ser9) [71]. Girk2C and Girk3 possess identical C-terminal PDZ interaction motifs (-ESKV). (b) Structure of the Girk-Gβγ complex from a side-view, with colors highlighting Girk2 (blue), Gβ (red), Gγ (green, with associated geranyl-geranyl (gg) lipid modification), PIP2 (yellow/orange sticks), Na+ ions (purple spheres), K+ ions (green spheres within the pore). Note that there are 4 Gβγ binding sites per channel, and the Gβγ facing the viewer has been removed for clarity [16].
