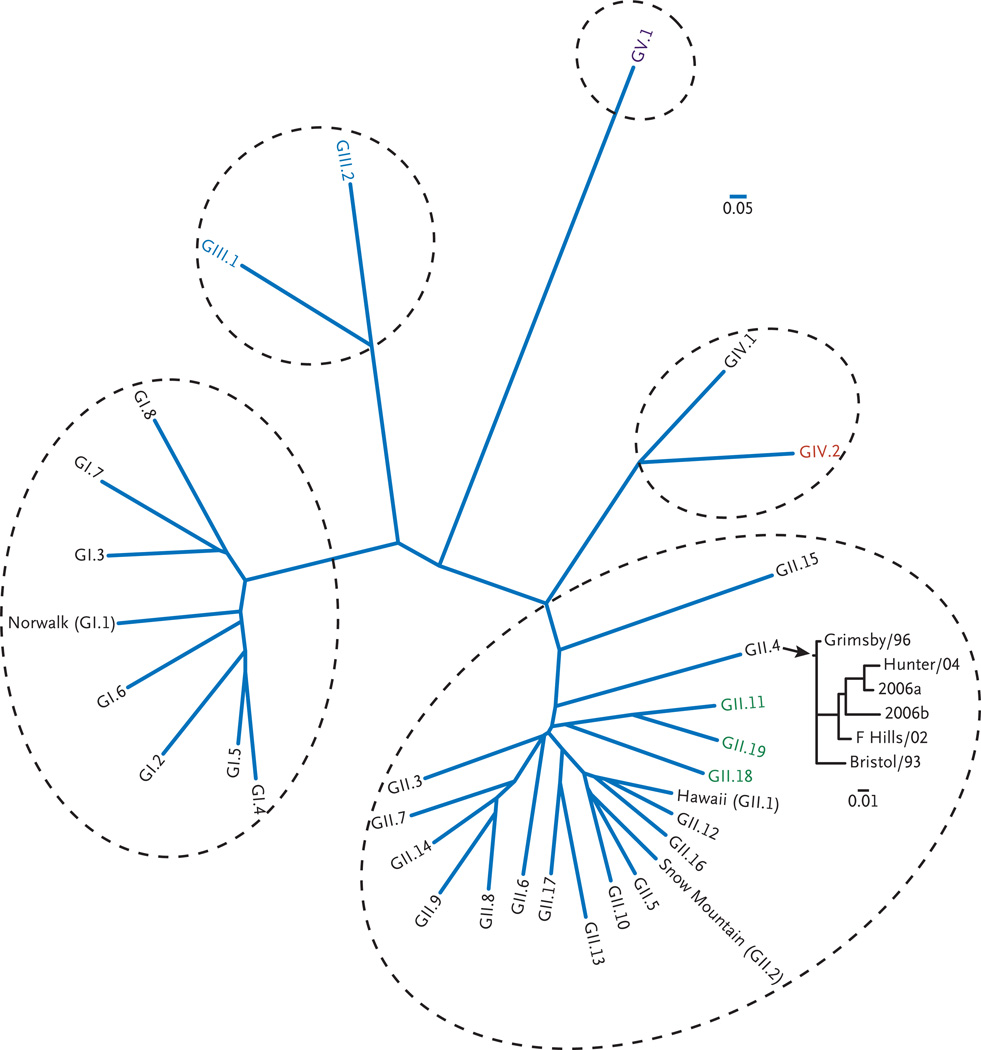
Figure 1. Phylogenetic Analysis of Noroviruses.
Noroviruses are a separate genus in the family Caliciviridae and have great diversity of genogroups, genotypes, and subtypes. Genogroups III and V have been identified only in animals. Strain GI.1 was the original Norwalk virus; other classic viruses named for the locations of outbreaks they caused are shown; strain GII.4 has become the predominant strain in the United States and throughout the world. This multiple alignment of 52 calicivirus viral protein (VP) 1 capsid amino acid sequences was performed with the use of Clustalw2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html), and the phylogenetic analyses were performed with programs in the Phylip package, version 3.6. The scale bars represent the unit for the expected number of substitutions per site. Similar analyses that were performed for recent GII.4 norovirus strains show the emergence of strains every 2 to 4 years. Human prototype viruses are listed in black, porcine viruses GII.11, GII.19, and GII.18 are shown in green, bovine viruses are shown in blue, a murine virus is shown in purple, and a lion virus GIV.2 is shown in red. The prototype strains and the sequence accession references used for this analysis are listed in Table 1 in the Supplementary Appendix.