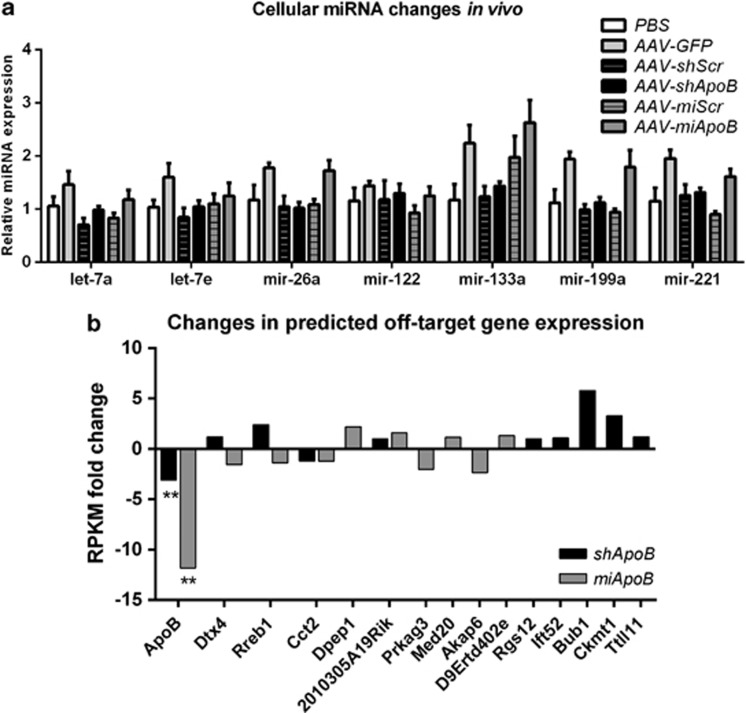
Figure 3.
Effect of shApoB and miApoB expression on cellular miRNA and off-target gene expressions. (a) Relative expression of let-7a, let-7e, mir-26a, mir-122, mir-133a, mir-199b and mir-221 in vivo was measured by miRNA Taqman. Mice were intravenously injected with 1 × 1011 gc AAV-shApoB or AAV-miApoB, or their respective controls AAV-shScr, AAV-miScr, AAV-GFP and PBS. Total RNA was isolated from snap-frozen livers. miRNA expression levels were calculated relative to actin mRNA and the PBS-treated group was set at 100%. Data are represented as mean +s.e., treatment groups are n=5. (b) Changes in predicted shApoB and miApoB off-target gene expression. Smith–Waterman algorithm was used to predict shApoB and miApoB off-target genes for the most abundant expressed variants of passenger and guide strands in mouse reference genome (15 May 2012 NCBI build 38.1). Predicted targets were manually screened for genes expressed in the liver that were detected in NGS transcriptome analysis. Gray and black bars represent fold change in AAV-shApoB and AAV-miApoB relative to AAV-shScr or AAV-miScr controls respectively. Kal's test (Z-test) with Bonferroni correction was used to compare the RPKM value in the shApoB versus shScr or miApoB versus miScr. **P<0.01.