Abstract
The NF1 tumor suppressor protein, neurofibromin, is a negative regulator of Ras. Neurofibromin is dynamically regulated by the proteasome and its degradation and re-expression are essential for maintaining appropriate levels of Ras-GTP. Like p53, NF1/neurofibromin can be inactivated in cancer by both mutations and excessive proteasomal destruction; however, little is known about the mechanisms that underlie this latter process. Here we show that a Cullin 3 (Cul3)/KBTBD7 complex controls both the regulated proteasomal degradation of neurofibromin and the pathogenic destabilization of neurofibromin in glioblastomas. Importantly RNAi-mediated Cul3 ablation and a dominant-negative Cul3 directly stabilize neurofibromin, suppress Ras and ERK, and inhibit proliferation in an NF1-dependent manner. Moreover, in glioblastomas where neurofibromin is chronically destabilized, Cul3 inhibition re-stabilizes the protein and suppresses tumor development. Collectively these studies demonstrate a previously unrecognized role for Cul3 in regulating Ras and provide a molecular framework that can be exploited to develop potential cancer therapies.
Keywords: Ras, NF1, neurofibromin, Glioblastoma multiforme, Cul3
INTRODUCTION
The Ras pathway is one of the most commonly deregulated pathways in human cancer(1–3). Oncogenic mutations in RAS genes are found in a diverse set of human tumors; however, Ras can also become hyperactivated as a consequence of loss-of-function mutations in genes that encode Ras GTPase Activating Proteins (RasGAPs), which negatively regulate Ras by catalyzing the hydrolysis of Ras-GTP (4–7). The NF1 tumor suppressor is the most well-studied RasGAP and is mutated in neurofibromatosis type I (NF1), a familial cancer syndrome affecting 1 in 3500 individuals world-wide(8). NF1 has also been shown to be mutated or lost in sporadic glioblastoma (9–11), non-small cell lung cancers(12), neuroblastoma (13,14), and melanoma(15–17), demonstrating a broader role for NF1in human cancer.
While tumor suppressors are commonly inactivated by genetic mechanisms, the proteasomal destruction of tumor suppressor proteins such as p53, p27, and PTEN also contributes to their functional inactivation in human cancer(18–21). Moreover, in tumor types where p53 is destabilized, strategies aimed at blocking its pathogenic destruction have been an active area of therapeutic development(22). Neurofibromin is also controlled by the ubiquitin-proteasome system and its expression is dynamically regulated by growth factors in normal settings(11,23). More recently, we and others have found that NF1/neurofibromin expression can be ablated by genetic as well as proteasomal mechanisms in sporadic glioblastomas(9–11). However, little is known about the molecular mechanisms that control its regulated or aberrant destruction. Here we identify the ubiquitin ligase complex that controls neurofibromin stability in normal and pathogenic settings. Thus, in addition to identifying a new regulator of the Ras pathway, these findings can be used as a foundation for developing new therapeutic approaches to treat cancers such as glioblastomas where neurofibromin is chronically destabilized. These findings also reveal a new strategy to suppress Ras activation, which may be more broadly exploited to develop therapies for other cancers.
RESULTS
Identifying the ubiquitin ligase complex that regulates neurofibromin degradation in response to growth factors
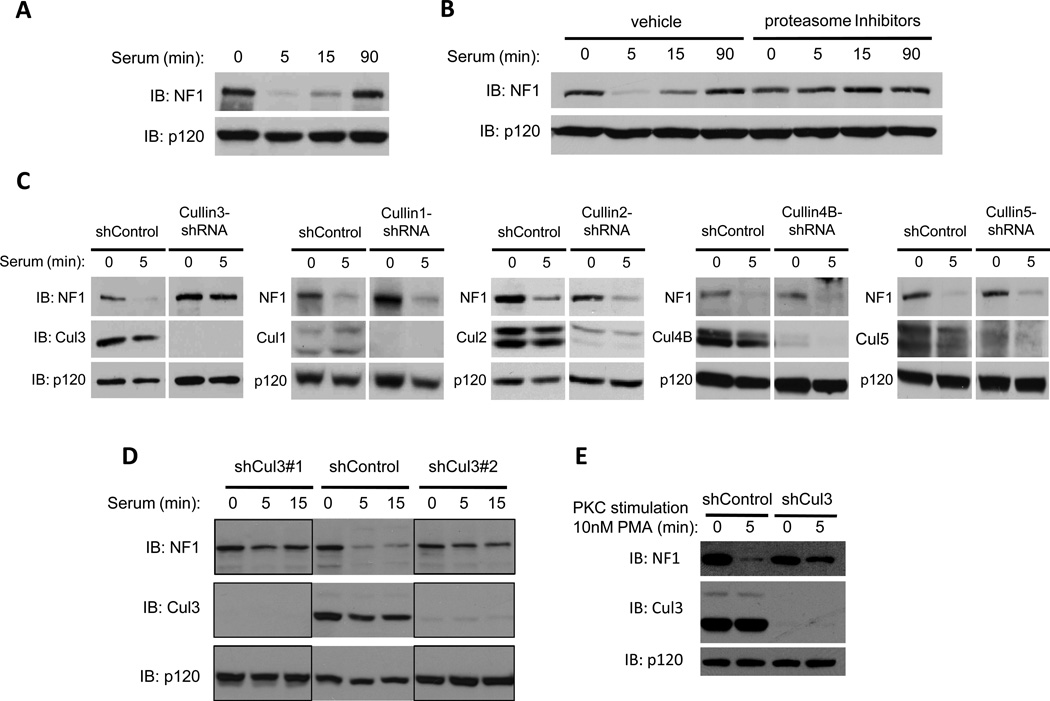
We previously reported that a variety of growth factors trigger the acute destruction of neurofibromin within 5 minutes of exposure, and that protein levels are re-elevated 90 minutes thereafter(23). Importantly, we showed that 1) this tightly regulated degradation and re-expression of neurofibromin controls both the amplitude and duration of Ras-ERK signaling in response to growth factors, 2) neurofibromin destruction is caused by ubiquitin-mediated proteasomal degradation, and 3) neurofibromin stabilization prevents aberrant cellular proliferation(23). The kinetics of neurofibromin degradation in response to serum are depicted in Fig. 1A and, as previously observed, its destruction can be blocked by proteasome inhibitors (Fig. 1B and(11,23). In contrast other RasGAPs, such as RasGAPp120, are not regulated by the proteasome (Fig. 1B).
Figure 1. Cul3 is required for the proteasomal degradation of neurofibromin.
A) Immunoblot indicating neurofibromin (NF1) degradation in serum-starved NIH3T3 fibroblasts stimulated with 10% serum for the times shown. RasGAPp120 (p120) is not degraded under these conditions and is shown as a loading control.
B) Serum-starved mouse embryonic fibroblasts (MEFs) were treated with a combination of proteasome inhibitors (1uM bortezomib, 10mM MG132) or vehicle (DMSO) for 2h and then stimulated with 10% serum for the times indicated. Immunoblots of neurofibromin and RasGAPp120 are shown.
C) NIH3T3 fibroblasts were infected with a lentiviral control or a lentivirus containing an shRNA sequence directed against specific mammalian cullin genes as noted. Knock-down of cullins was confirmed by immunoblot (IB). Neurofibromin and RasGAPp120 immunoblots of serum-starved cells stimulated with 10% serum for the times indicated are shown.
D) Immunoblot of neurofibromin degradation in NIH3T3 fibroblasts expressing a control shRNA vector or distinct shRNA constructs targeting different regions of the Cul3 transcript. Cul3 depletion is confirmed by immunoblot, and RasGAPp120 levels are shown as a loading control.
E) Immunoblot of neurofibromin degradation in control or shCul3-expressing NIH3T3 fibroblasts following PKC activation (10nM PMA) for 5 min. Cul3 depletion and a RasGAPp120 loading control are also shown.
To identify the ubiquitin ligase responsible for neurofibromin degradation, we utilized an shRNA-based screening approach. We focused on the cullin-RING superfamily of E3 ligases, as these proteins have been shown to play a critical role in cell cycle regulation and growth control(24,25). Cells were infected with lentiviruses encoding shRNA sequences that recognize each of the mammalian cullin subunits. Depletion of Cullin 3 (Cul3), but not other cullins, blocked the degradation of neurofibromin in response to serum (Fig. 1C). Notably, Cullin 1 (Cul1), which controls the stability of potential oncogenes and tumor suppressor proteins involved in cell cycle progression and proliferation such as Cyclin E and p27(26), had no effect on neurofibromin degradation (Fig. 1C). Two distinct shRNA constructs targeting different regions of Cul3 blocked neurofibromin degradation, demonstrating that this phenotype is not an off-target effect (Fig 1D).
We have previously shown that growth factors trigger neurofibromin destabilization by activating protein kinase C (PKC), which provides a signal that is both necessary and sufficient for neurofibromin destruction(11). To determine whether Cul3 mediates PKC-regulated neurofibromin degradation, cells were treated with the PKC activator PMA (Phorbol-12-Myristate-13-Acetate). Whereas neurofibromin was degraded in response to PKC activation in control cells, PMA-induced degradation was blocked in Cul3-depleted cells (Fig. 1E). These results indicate that Cul3 mediates the PKC-driven destabilization of neurofibromin.
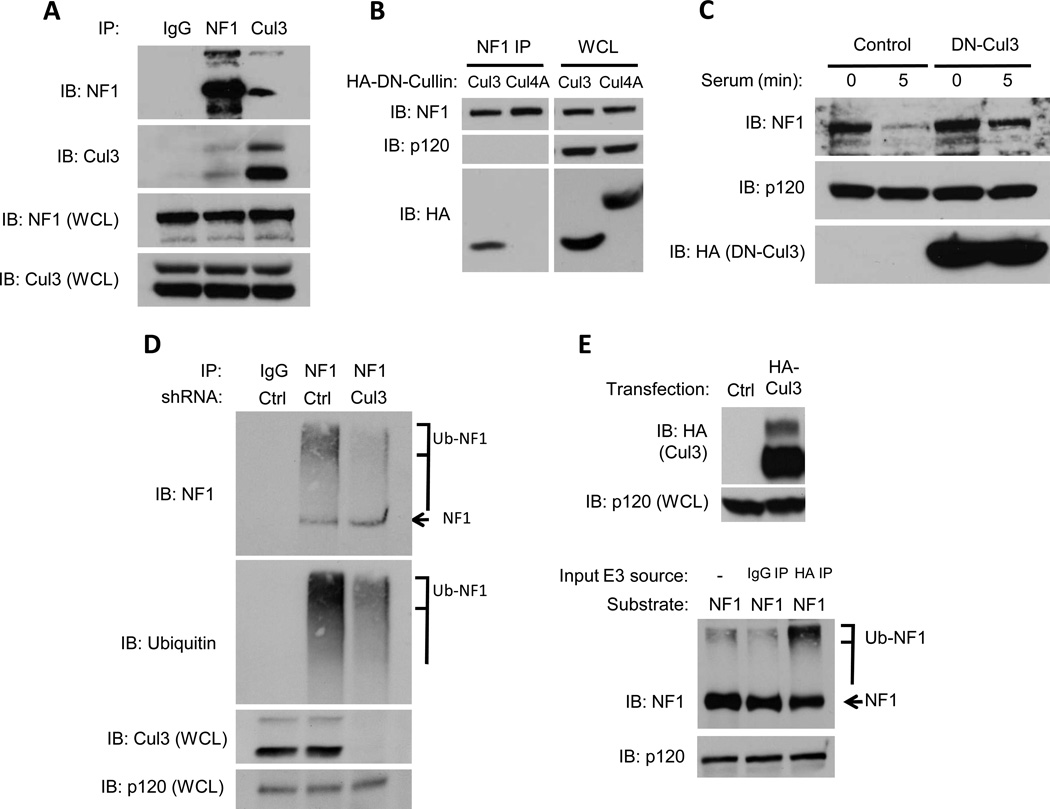
Endogenous Cul3 interacts with neurofibromin and controls neurofibromin ubiquitination and degradation
To determine whether Cul3 directly regulates neurofibromin stability cells were pre-treated with proteasome inhibitors and neurofibromin or Cul3 were immunoprecipitated from serum-stimulated cells. Importantly, endogenous neurofibromin and endogenous Cul3 co-precipitate under the precise conditions in which neurofibromin is normally degraded(Fig. 2A). To confirm a functional role for Cul3 in this complex we utilized a dominant-negative fragment of Cul3 (DN-Cul3). When expressed as a truncated fragment, the N-terminal half of Cul3 functions as a dominant negative protein because it can bind Cul3 targets but is unable to recruit the E2 ubiquitin-conjugating enzyme(27,28). As such, Cul3 substrates directly associate with this fragment but cannot become ubiquitinated or degraded. Similar to endogenous Cul3, DN-Cul3 also formed a stable complex with endogenous neurofibrominin vivo (Fig. 2B). Neurofibromin did not co-precipitate with a DN-Cul4A fragment, further highlighting the specificity of this interaction. Similar to the effects of Cul3 shRNAs, DN-Cul3 substantially blocked neurofibromin degradation (Fig. 2C).
Figure 2. Cul3 associates with neurofibromin and regulates its ubiquitination.
A) Endogenous neurofibromin, Cul3, or a control were immunoprecipitated from serum-starved NIH3T3 fibroblasts after stimulation with 10% serum in the presence of proteasome inhibitors and a chemical cross-linker. Immunoblots of neurofibromin or Cul3 immunocomplexes and whole cell lysates (WCL) are shown.
B) Neurofibromin was immunoprecipitated from NIH3T3 fibroblasts expressing dominant-negative (DN) versions of Cul3 and Cul4A. Expression of DN cullins in total cell lysates was confirmed with an anti-HA antibody and equal loading was confirmed by a RasGAPp120 immunoblot. Immunoblots using a neurofibromin antibody and an HA antibody confirm the specific association of DN-Cul3 with neurofibromin.
C) Immunoblot assessing the effect of lentivirally-expressed HA-tagged DN-Cul3 or a lentiviral control vector on neurofibromin stability in serum-stimulated NIH-3T3 fibroblasts. Expression of DN-Cul3 is detected using an anti-HA antibody. RasGAPp120 levels are shown as a loading control.
D) Serum-starved NIH3T3 fibroblasts expressing a control lentiviral vector or a Cul3-specific shRNA were treated with proteasome inhibitors (1uM bortezomib, 10mM MG132) for 2 h, and stimulated with 10% serum for 5 minutes. Total cell lysates were isolated in buffer containing 2nM NEM to preserve polyubiquitin chains. Neurofibromin or control immunoprecipitations are shown. Both ubiquitin and neurofibromin blots denote a decrease in high-mobility ubiquitinated neurofibromin (brackets) in cells depleted of Cul3. An arrow points to unmodified neurofibromin. Cul3 levels and a RasGAPp120 loading control are also shown.
E) (Top) Western blot depicting the expression of ectopic HA-tagged Cul3 used for in vitro ubiquitination reactions. (Bottom) Neurofibromin blot demonstrating an increase in ubiquitinated neurofibromin catalyzed by endogenous Cul3 complexes immunoprecipiated from stimulated cells. These in vitro reactions included recombinant human E1, E2 (UBCH5a), ubiquitin and an ATP regenerating system. An arrow points to unmodified neurofibromin, and a bracket indicates the location of polyubiquitinated neurofibromin.
Proteins that are targeted to the proteasome for degradation are first covalently modified with polyubiquitin chains resulting in the accumulation of higher-mobility species of the target protein(29). To confirm that Cul3 indeed controls ubiquitination, we examined the ubiquitination state of neurofibromin in wild type cells or in cells in which Cul3 expression was suppressed by RNAi. Consistent with previous reports, neurofibromin is ubiquitinated in serum-treated cells exposed to proteasome inhibitors, which can be visualized as a high mobility smear that can be detected by both NF1 and ubiquitin antibodies(Fig. 2D)(11,23). Importantly, Cul3-specific shRNA sequences dramatically suppressed neurofibromin ubiquitination (Fig. 2D). To determine whether Cul3 was directly mediating this ubiquitination we generated a Cul3 expression construct and confirmed that immunoprecipitated, recombinant Cul3 could ubiquitinate Nrf2, a well-established Cul3 substrate (Fig. 2E and Supp. Fig. 1). This immunopurified, recombinant Cul3 also ubiquitinated neurofibromin in a reconstituted in vitro ubiquitination assay (Fig. 2E). It should be noted that while ubiquitinated neurofibromin extracted from cells is manifested as a high mobility smear on immunoblots, we have consistently found that neurofibromin ubiquitinated in vitro migrates as amore concentrated high mobility species, with less of a smeared pattern(11,23). This is likely due to the large size of ubiquitinated neurofibromin (250 kDa), which becomes maximally ubiquitinated in vitro, in addition to the activity of de-ubiquitinating enzymesin vivo. Together, the endogenous co-precipitation data, RNAi and dominant-negative experiments, as well as thesein vitro and in vivoubiquitination studies demonstrate that Cul3 directly controls the dynamic ubiquitination and degradation of neurofibromin.
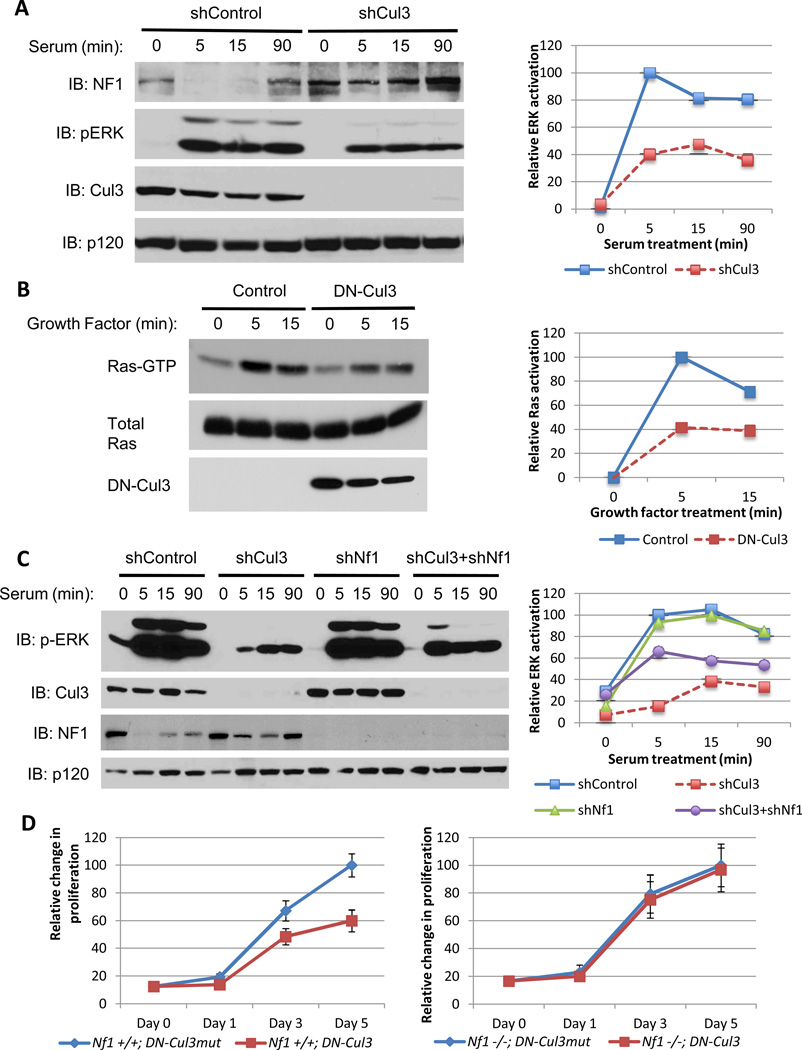
Cul3 loss suppresses Ras/ERK signaling
Because neurofibromin is a RasGAP the effects of Cul3 suppression on the Ras/ERK pathway were examined. In response to serum and growth factors ERK and Ras are maximally activated within 5 minutes (Fig. 3A, B). However Cul3-specific shRNAs suppressed ERK activity by nearly 60% (Fig. 3A). DN-Cul3 exhibited a similar suppressive effect (60%) on Ras-GTP levels directly (Fig. 3B) and similarly suppressed ERK activity (Supp. Fig. 2A). We next evaluated the relative contribution of neurofibromin stabilization in mediating this suppressive effect on Ras signaling. Cul3 shRNA constructs suppressed ERK activation by 87% (Fig. 3C), underscoring the potent effects of Cul3 suppression on this pathway. RNAi-mediated neurofibromin suppression had no effect on ERK activity on its own in this setting (Fig. 3C). However,Nf1-ablation substantially rescued the defects in ERK signaling, increasing ERK activation 6-fold and restoring it to 66% of maximal levels (Fig. 3C). Taken together these data demonstrate that Cul3-mediated neurofibromin degradation plays an essential role in regulating Ras and ERK and that Cul3 suppression potently inhibits this pathway. Surprisingly however, while depletion of neurofibromin substantially restored ERK activation in Cul3-deficient cells, a complete rescue was not observed, suggesting that Cul3 may have additional targets that impact Ras activation. Thus, in addition to revealing a previously unrecognized role for Cul3 in regulating Ras, the observation that Cul3 may affect this pathway through neurofibromin and possibly an additional target protein, strengthens the potential utility of Cul3 inhibition in suppressing the Ras pathway.
Figure 3. Cul3 loss suppresses Ras/ERK signaling and proliferation.
A) Left, immunoblot of neurofibromin, phosphorylated ERK, Cul3 and RasGAPp120 from serum-stimulated NIH3T3 fibroblasts expressing a lentiviral control shRNA or a Cul3-specific shRNA. Right, quantification of relative levels of activated ERK in shControl or shCul3-expressing cells using image j software.
B) Left, immunoblot of a Ras-GTP pulldown assay to assess activation of Ras in mouse embryonic fibroblasts expressing a lentiviral control vector or dominant-negative Cul3 (DN-Cul3) following treatment with LPA for the times indicated. Right, relative levels of Ras activation quantified using image j software and depicted as a percent of maximal Ras activation compared to vector control-expressing cells.
C) Left, immunoblot of phosphorylated ERK, Cul3, neurofibromin, and RasGAPp120 from NIH3T3 fibroblasts expressing lentiviral control vectors or Cul3 and NF1-specific shRNAs individually or in tandem. Right, quantification of relative activated ERK levels using image j software, denoting that the attenuation in ERK activation due to Cul3 deficiency is rescued in part by the ablation of Nf1 expression.
D) Relative proliferation curves of NF1 wild-type (Nf1+/+) or NF1-null (Nf1−/−) mouse embryonic-fibroblasts expressing a dominant-negative Cul3 (DN-Cul3) or an inert mutant form of this construct (DN-Cul3mut) as a control. Error bars represent the standard deviation (SD) of triplicate cell number measurements.
Growth suppression conferred by dominant-negative Cul3 is dependent on NF1
The amplitude and duration of Ras/ERK activation have been shown to play an important role in restricting or permitting cellular proliferation(30). We therefore evaluated the effects of Cul3 suppression on cellular proliferation. shRNA-mediated depletion of Cul3 in mouse embryonic fibroblasts (MEFs) led to acute cell death (not shown), indicating that a minimal threshold of Cul3 activity is required for the viability of these cells, as has been previously reported(31). However, expression of dominant-negative Cul3 (DN-Cul3)was tolerated. To control for the effects of DN-Cul3 in these cells, we also evaluated a mutant form of DN-Cul3 that cannot bind its substrates (L52AE55A), hereafter referred to as DN-Cul3mut (32). We validated these DN-Cul3 constructs by showing that expression of DN-Cul3 stabilized the transcription factor Nrf2, a well-established target of Cul3, whereas expression of DN-Cul3mut had no effect (Supp. Fig. 2B). Importantly, DN-Cul3 but not DN-Cul3mut inhibited proliferation and ultimately caused a growth arrest in wild-type MEFs (Fig. 3D). In contrast, Nf1−/− MEFs, which lack neurofibromin, were unaffected by the expression of DN-Cul3 and proliferated indistinguishably fromNf1−/− MEFs expressing DN-Cul3mut(Fig. 3D). Notably, DN-Cul3 but not DN-Cul3mut led to a decrease in activated ERK in wild-type MEFs, whereas ERK activation was not hampered by DN-Cul3 in Nf1−/− MEFs (Supp. Fig. 2A). These results demonstrate that the growth inhibitory effects of Cul3 suppression are dependent on neurofibromin.
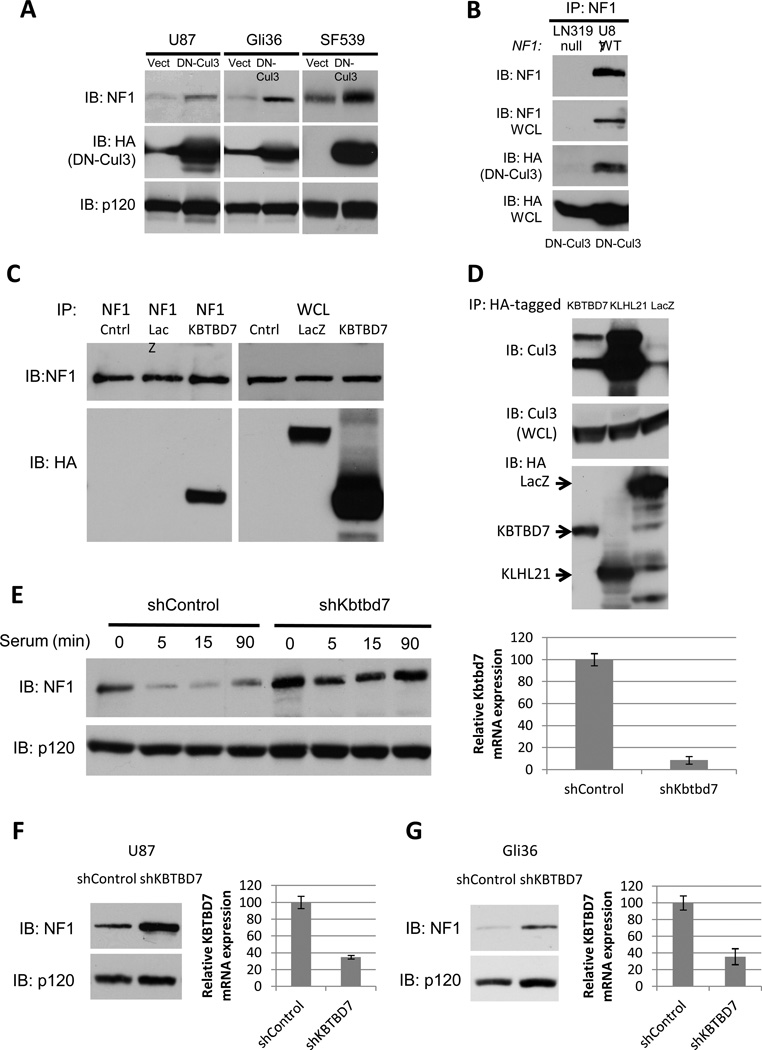
Cul3 regulates neurofibromin destabilization in GBMs
We and others have shown that one NF1 allele is mutated or lost in 15–23% of sporadic glioblastomas. However biallelic inactivation only occurs in 3% of tumors and cell lines(9–11,33). Nevertheless, neurofibromin protein expression is suppressed or absent in a much higher fraction of tumors(11). We have shown this suppression is caused by excessive ubiquitin-mediated proteasomal degradation and that neurofibromin destabilization plays an active role driving tumor pathogenesis(11). To determine whether Cul3 was responsible for the destabilization of neurofibromin in GBMs we expressed DN-Cul3 in three cell lines where neurofibromin has been shown to be destabilized: U87MG, Gli36 and SF539. In all cases DN-Cul3 expression resulted in an increase in the expression of neurofibromin protein (Fig. 4A). Cul3 shRNA sequences similarly stabilized neurofibromin in these cells (Supp. Fig. 3). Importantly, DN-Cul3 also co-precipitated with endogenous neurofibromin (Fig. 4B), suggesting that these effects were direct, consistent with the analysis shown in Fig. 2.
Figure 4. Cul3 and the adaptor protein KBTBD7 destabilize neurofibromin in GBM cells.
A) Immunoblot of neurofibromin, RasGAPp120 and HA-tagged dominant-negative Cul3 (DN-Cul3) from GBM cell lines expressing lentiviral HA-DN-Cul3 or a vector control.
B) Immunoblot of neurofibromin IP from NF1-wild type (U87) or NF1-null (LN319) GBM cells expressing HA-DN-Cul3. HA blot indicates that HA-DN-Cul3 associates with neurofibromin in U87 cells.
C) Immunoblot of neurofibromin IP from proteasome-inhibited, serum-stimulated cells expressing HA-KBTBD7 or HA-LacZ. HA blot indicates that KBTBD7 associates specifically with neurofibromin.
D) Immunoblot of endogenous Cul3 co-precipitating with HA-tagged BTB proteins KBTBD7 and KLHL21 expressed in 293T cells. Total Cul3 levels are shown as a loading control.
E) Left, immunoblot of neurofibromin degradation in serum-starved NIH3T3 fibroblasts stimulated with 10% serum for the times shown in the absence or presence of a short hairpin RNA specific for Kbtbd7. RasGAPp120 is shown as a loading control. Right, relative expression of Kbtbd7 mRNA as assessed by normalized quantitative PCR.
F) Left, immunoblot of neurofibromin in U87 GBM cells expressing a control or KBTBD7-specific shRNAs. RasGAPp120 is shown as a loading control. Right, relative expression of KBTBD7 mRNA as assessed by normalized quantitative PCR.
G) Left, immunoblot of neurofibromin in control Gli36 GBM cells or cells expressing a KBTBD7-specific shRNA. RasGAPp120 is shown as a loading control. Right, relative expression of KBTBD7 mRNA as assessed by normalized quantitative PCR.
The Cul3 adaptor protein KBTBD7 regulates neurofibromin stability
Cul3, like all cullins, recruits its targets through a discrete set of substrate recognition adaptor proteins. Cul3 substrate adaptors contain a BTB protein-protein interaction domain (for Bric-a-brac, Tramtrack, Broad complex)(27,32,34,35). BTB-domain proteins bind to Cul3 through the BTB domain, and recruit substrates through diverse protein-protein interaction motifs(36). To further dissect the mechanism by which Cul3 regulates neurofibromin stability, we performed a proteomic/mass-spectrometry-based screen to identify the neurofibromin-associated BTB adaptor protein. Similar strategies have been used to identify components of E3 ubiquitin ligase complexes for a variety of substrates(37–39). Specifically, mass spectrometry was performed on endogenous neurofibromin immunoprecipitates, generated from serum-stimulated cells in the presence of proteasome inhibitors. Using this approach, we detected the BTB domain-containing protein, KBTBD7, in the neurofibromin immunoprecipitates. To validate this interaction we expressed an epitope tagged KBTBD7 construct, as reliable antibodies for KBTBD7 are not presently available. Importantly, HA-KBTBD7 could be readily detected in a complex with endogenous neurofibromin (Fig. 4C). While KBTBD7 contains a BTB domain, its ability to interact with Cul3 has not been previously evaluated. Importantly, endogenous Cul3 co-precipitated with KLHL21, an established Cul3-binding BTB protein (40)as well as KBTBD7 (Fig. 4D). Moreover shRNA-mediated depletion of KBTBD7 blocked the serum-stimulated degradation of neurofibromin in NIH3T3 cells (Fig. 4E), mirroring the block in neurofibromin degradation caused by Cul3 depletion (Fig. 1,2). Finally, RNAi-mediated KBTBD7 depletion caused an increase in neurofibromin levels in GBM cell lines in which neurofibromin is normally destabilized (Fig. 4F, G). Together, these findings demonstrate that Cul3 utilizes KBTBD7 as the adaptor protein to regulate neurofibromin stability.
DN-Cul3 suppresses transformation and tumor growth through its effects on neurofibromin
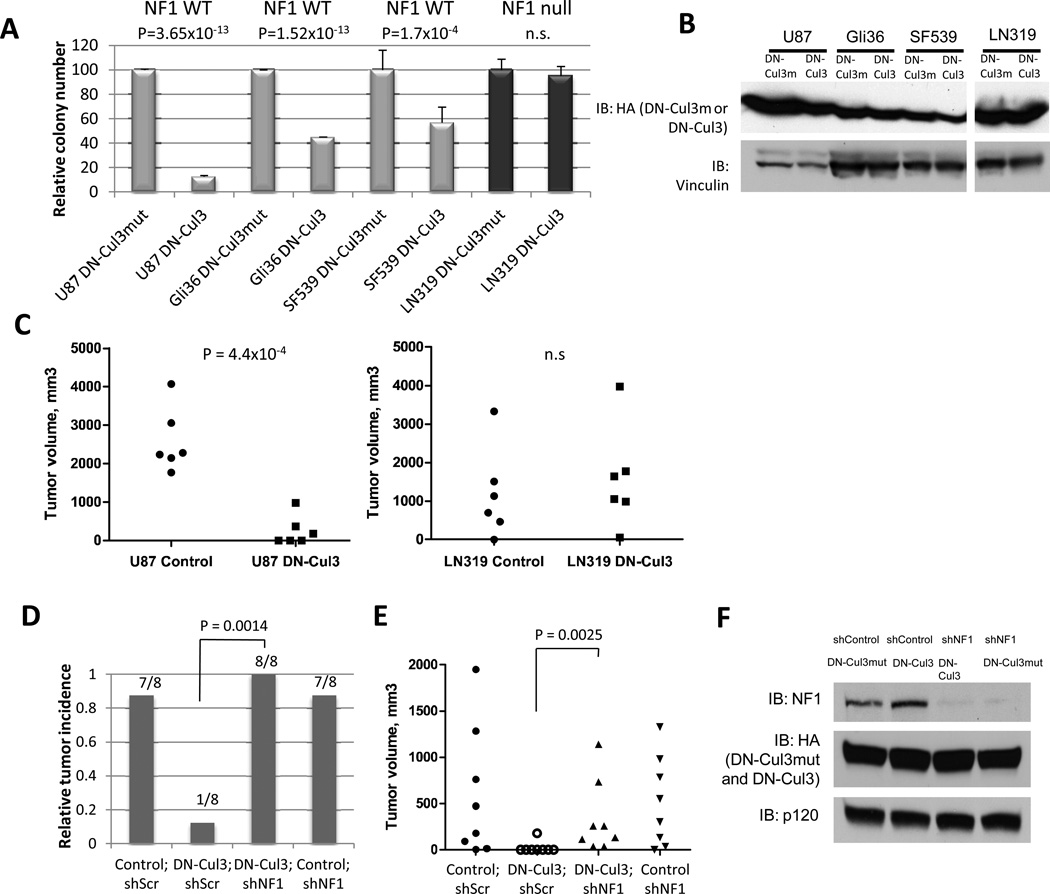
The observation that neurofibromin was stabilized by Cul3 shRNAs and DN-Cul3 raised the intriguing possibility that Cul3 inhibition might also suppress the transformed properties of these cancer cells. To ascertain the biological consequences of Cul3 suppression in GBMs, we compared the effects of the DN-Cul3 in GBM cells in which neurofibromin was destabilized by the proteasome, as compared to cells that were confirmed to be genetically NF1-deficient(11). Expression of DN-Cul3 significantly reduced the ability of NF1 wild-type/neurofibromin destabilized GBM cells to form colonies in soft agar as compared to cells that expressed equivalent levels of the control DN-Cul3mut, which had no effect preventing colony growth (Fig. 5A, B). In contrast, DN-Cul3 had no effect on the ability of NF1-null cells to form colonies (Fig 5A,B).
Figure 5. DN-Cul3 suppresses transformation and tumor growth through its effects on neurofibromin.
A) Relative differences in soft agar colony growth of NF1-WT (U87, Gli36, SF539) or NF1-null (LN319) GBM cells expressing DN-Cul3 or an inert mutant (L52AE55A)control (DN-Cul3mut). Data are represented as mean ± SD of triplicate measurements. P-values shown in the panel were calculated by Student’s t-test. n.s.: not statistically significant.
B) Immunoblot showing equivalent loading of DN-Cul3 and DN-Cul3mut in each GBM cell line used for soft-growth agar assays and xenograft injections shown in Fig. 5A. and Fig. 5C. Vinculin levels are shown as loading control.
C) Dot plot depicting differences in tumor volume in xenograft tumor-bearing mice injected with U87 or LN319 cells expressing DN-Cul3 or a (L52AE55A)DN-Cul3mut control (same cells as in A). P-value significance in the panel was determined by Student’s t-test. n = 6 each. n.s.: not statistically significant.
D) Relative tumor incidence of U87-shp53 GBM cells expressing DN-Cul3 or a control DN-Cul3mut (L52AE55A)vector in conjunction with a lentiviral NF1 shRNA or a control shRNA. Bars represent the fraction of tumor incidence over a period of 5 months and the number of tumors formed/number of injections are depicted. n = 8 in each category. P = 0.0014 (DN-Cul3;shScr versus DN-Cul3;shNF1) using Fisher’s exact test.
E) Dot plot depicting tumor volume in xenograft-tumor bearing mice injected with U87-shp53 GBM cells expressing DN-Cul3 or the control DN-Cul3mut (L52AE55A)vector together with a lentiviral NF1 shRNA or a control shRNA. A Mann-Whitney U test (P = 0.0025) was used to illustrate a significant difference in tumor volume in xenograft tumors comprised of DN-Cul3;shScr versus those comprising DN-Cul3;shNF1-expressing cells.
F) Neurofibromin expression in shp53-U87 cells used for xenograft studies shown in Fig. 5D and Fig. 5E. Cells express shNF1 or a control shRNA and HA-tagged DN-Cul3 or DN-Cul3mut constructs. HA immunoblot showing equivalent expression of DN-Cul3 and DN-Cul3mut is shown, as well as RasGAPp120 which serves as a loading control.
Finally, we investigated whether DN-Cul3 activity could affect the ability of GBM cells to form xenograft tumors in vivo. While U87 cells readily formed tumors in the flanks of immunocompromised mice, DN-Cul3 potently suppressed tumor development (Fig. 5C)(P=0.00044). In contrast, expression of DN-Cul3 did not prevent tumor growth in mice injected withNF1-null LN319 cells, which are genetically NF1-deficient. Because DN-Cul3 likely affects other targets, we directly evaluated the contribution of neurofibromin stabilization in tumor suppression. To examine this in an isogenic setting we utilized U87shp53 cells, which are able to tolerate complete NF1 ablation(11). DN-Cul3 but not control DN-Cul3mut similarly inhibited tumor growth when expressed in these cells. However, RNAi-mediated NF1 suppression restored tumorigenicity (Fig. 5D–F), indicating that the tumor suppressive effects of DN-Cul3 were dependent on NF1. Collectively, these data demonstrate that Cul3 suppression can potently inhibit the tumorigenic properties of glioblastomas in which neurofibromin has been inactivated by the proteasome, thus revealing a mechanism-based vulnerability that can be used to develop new potential therapies. Importantly, this vulnerability may extend beyond glioblastomas and may ultimately be harnessed as a means of suppressing Ras signaling in other cancers. A model describing the pathway identified here is shown in Figure 6.
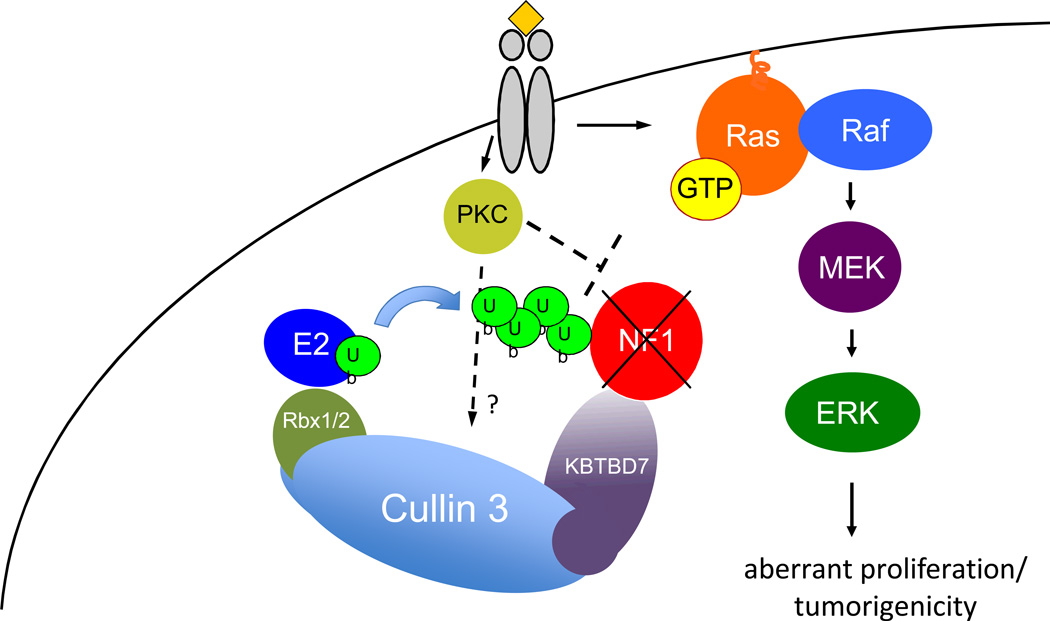
Figure 6. The Cul3 E3 ligase regulates NF1 ubiquitination and stability in normal and pathogenic settings.
Upon growth factor receptor stimulation PKC becomes activated which triggers the ubiquitination and degradation of neurofibromin by the proteasome. This ubiquitination is catalyzed by the Cul3 E3 ligase and the BTB adaptor protein KBTBD7. In normal cells this degradation contributes to Ras/ERK activation. Shortly thereafter neurofibromin levels become re-elevated and properly terminate the Ras signal. However excessive PKC activity in some GBMs results in chronic neurofibromin destabilization. The data presented here also demonstrate that Cul3 and KBTBD7 also mediate this pathogenic destruction. NF1: Neurofibromin; PKC: Protein Kinase C; Ub: ubiquitin; E2: ubiquitin-conjugating enzyme; Rbx1/2: RING-box protein1/2; KBTBD7: kelch-repeat and BTB-containing protein 7.
DISCUSSION
The Ras pathway plays an essential role in transducing signals from activated growth factor receptors and regulates a wide variety of biological responses(30). The proper intensity of Ras/ERK signaling has been shown to be essential for dictating specific cellular responses and can determine whether a cell proliferates, arrests, differentiates, survives or dies(30,41). However, there is still little known about the precise molecular events that regulate the timing, duration, and intensity of Ras signaling that control these diverse biological behaviors. Here, we report that Cul3 and the BTB adaptor protein KBTBD7 regulate the ubiquitination and rapid degradation of neurofibromin in response to growth factors. Although we cannot rule out the involvement of additional ligases in some settings, in this report we show that Cul3 is essential for full activation of the Ras/ERK pathway. Moreover, Cul3 suppression potently inhibits Ras/ERK activation and cellular proliferation through its effects on neurofibromin. Thus, these studies reveal a previously unrecognized regulatory component of the Ras signal transduction pathway and provide fundamental insight into how Ras activity is so exquisitely regulated.
Notably, the excessive destabilization of neurofibromin has also been shown to underlie the pathogenesis of glioblastomas(11). Here we show that Cul3 and KBTBD7 also mediate this instability. Moreover, Cul3 suppression potently inhibits tumorigenicity by stabilizing neurofibromin. We have previously reported that PKC activation triggers neurofibromin degradation in response to growth factors and that excessive PKC activity causes chronic destabilization in a subset of glioblastomas(11,23). The molecular events that drive this destruction are currently unknown; however, the insight provided in this study will serve as the foundation for future investigations. Importantly, an oncogenic role for Cullin 3 or KBTBD7 has not previously been described in the pathogenesis of human cancer. Moreover the involvement of Cul3 in regulating neurofibromin and Ras may be exploited to suppress tumor development. For example based on these studies a Cul3 or a general cullin inhibitor would be expected to inhibit tumors in which neurofibromin is destabilized (e.g. GBMs). Notably, a small molecule inhibitor that targets NEDD8, a critical regulatory component of the cullin-RING ligases, has already been developed (MLN4924)(42). MNL4924 has been shown to trigger cell death in a variety of cancer cell lines, exhibits significant antitumor activity in xenograft assays, and is presently being evaluated in several phase I clinical trials for solid tumors and hematological malignancies(42,43). More recently cell-based screens identifying inhibitors of individual cullin-RING ligases have been reported(44–46), describing the first attempts at drug discovery for this class of ubiquitin ligases. Finally, the insight that p53 is degraded by the ubiquitin-proteasome system has inspired the development of numerous small molecule inhibitors specifically aimed at blocking its destruction, which are now being extensively evaluated in the clinic(22,47). Thus the observations presented here should provide the molecular framework and rationale to evaluate and develop agents aimed at blocking neurofibromin destruction and Ras activation, which may impact therapeutic development in glioblastoma, and possibly other cancers which more generally rely on RTK/Ras signaling networks.
METHODS
Cell Culture
NIH3T3 fibroblasts were cultured in DMEM supplemented with calf serum. Primary mouse embryonic fibroblasts (MEFs) and 293T cells were cultured in DMEM supplemented with fetal calf serum and L-glutamine. For serum starvation involving NIH3T3 fibroblasts, cells were trypsinized, neutralized with 0.5 mg/ml soybean trypsin inhibitor in DMEM, washed and plated as indicated below in serum-free media. MEFs were made quiescent as previously described (48) by first growing to confluency over 72 hours, then changing media to 0.1% fetal calf serum for 72 additional hours and washed, resuspended and plated in serum-free media for 18 hours prior to stimulation. U87, Gli36, SF539 and LN319 GBM cells were cultured in DMEM supplemented with 10% fetal calf serum and L-glutamine. All non-MEF cell lines were obtained from the ATCC. No additional authentication was performed by the authors.
Degradation time courses and proteasome inhibition
NIH3T3 fibroblasts were plated in serum-free DMEM at a density of 5 × 105 cells/10-cm culture dish. After 18 hours, cells were stimulated with 10% calf serum for the times indicated. For PKC stimulation, cells were treated with 10 nM PMA. Quiescent MEFs were plated at a density of 5 × 105 cells/10-cm culture dish for 18 hours and stimulated with 10% fetal calf serum or 6uM LPA for the times indicated. Cells were lysed at specified time points with 1% SDS boiling lysis buffer and clarified. For experiments that required proteasome inhibition as indicated in the text, cells were pre-treated with 1uM bortezomib, (LC laboratories) and 10uM MG-132 (Boston Biochem) or vehicle (DMSO) for 2 hours prior to stimulation with serum or LPA.
Immunoblotting
Clarified cell lysates were normalized for protein concentration, separated by SDS-PAGE and transferred to Immobilon-P PVDF membrane for immunoblotting with the following antibodies: neurofibromin antibodies: UP69: a polyclonal antibody raised against a KLH-conjugated peptide, RNSIKKIV, 1:5000; or NF1-A300-140A (Bethyl), 1:1000; p120RasGAP (Transduction Laboratories), 1:2000; HA (for epitope-tagged contructs) (12CA5, Roche), 1:1000; Cul3 (BD-Transduction), 1:1000; pan-Ras (Millipore), 1:20,000; Nrf2 (C-20, Santa Cruz 1:1000), phosphorylated ERK (Transduction Laboratories), 1:3000; ERK (Transduction Laboratories, 1:3000); ubiquitin (DAKO, 1:1000)
Interactions between neurofibromin and Cul3
To detect interactions between endogenous Cul3 and neurofibromin, serum-starved NIH3T3 cells were treated with proteasome inhibitors and stimulated with serum, as described above. At the time of serum stimulation, the chemical cross-linker DSP (dithiobis[succinimidylpropionate]) (Thermo Scientific) was added to a final concentration of 1mM for 20 min, and the reaction quenched with 100mM Tris for 10 min, prior to lysis in ice-cold IP buffer (0.3% CHAPS, 10mM HEPES pH7.5, 120nM NaCl, 1mM EDTA, 10mM sodium pyrophosphate, 50mM sodium fluoride) supplemented with protease and phosphatase inhibitors (Roche). Lysates were incubated for 15-minutes on ice, clarified, combined and split into equivalent samples for parallel immunoprecipitations of neurofibromin, Cul3, or a control reaction with 2ug each of neurofibromin-D antibody (Santa Cruz), Cul3 antibody (H-293) (Santa Cruz), or normal rabbit IgG (Cell Signaling) rotating overnight at 4°C. Immunoprecipitations were immobilized on Protein A beads, rotating 1 h at 4°C, and washed 3x with IP buffer before denaturation and separation by SDS-PAGE. Western blots were probed with antibodies as described above, using conformation-specific secondary antibodies (Rockland). To detect interactions between DN-Cul3 and neurofibromin, cells were transduced with DN-Cul3 (see below) or control vectors as described in the text. Neurofibromin was immmunoprecipitated from cells using IP buffer supplemented with protease and phosphatase inhibitors only (Roche). Samples were immobilized on Protein A beads, washed 3x with IP buffer and separated by SDS-PAGE prior to Western blot.
Detection of ubiquitinated NF1 in vivo
NIH3T3 cells were stably transduced with a pLKO control vector or pLKO containing a Cul3 shRNA and plated in serum-free DMEM at a density of 5 × 105 cells/10-cm culture dish. After 18 hours, cells were treated with a combination of 10uM MG132 and 1uM bortezomib for 2hr, then stimulated for 5 min with 10% calf serum before lysing cells. To enrich for ubiquitinated neurofibromin, 3 10-cm dishes for each condition were washed and lysed in ice-cold RIPA buffer containing protease and phosphatase inhibitors (Roche), supplemented with 2mM N-ethyl-maleimide (NEM) (Sigma), which inactivates de-ubiquitinase enzymes. Lysates were clarified and precleared with Protein A beads, and neurofibromin was immunoprecipitated with 2ug neurofibromin-D antibody (Santa Cruz), and an equivalent control sample with rabbit IgG (Cell Signaling), rotating overnight at 4°C. Immunocomplexes were immobilized on Protein A beads, rotating 1 h at 4°C, then washed 3x with wash buffer prior to denaturation and separation by SDS-PAGE. Western blots were probed with ubiquitin antibody (DAKO), then stripped and reprobed with neurofibromin antibody (Bethyl), using conformation-specific secondary antibodies (Rockland).
In vitro ubiquitination
Neurofibromin immunoprecipitated from cycling NIH3T3 cells was used as a ubiquitination substrate (ATCC). Specifically, two 15-cm dishes were washed 3x with PBS and lysed with ice-cold IP buffer (0.3% CHAPS, 10mM HEPES pH7.5, 120nM NaCl, 1mM EDTA, 10mM sodium pyrophosphate, 50mM sodium fluoride) supplemented with protease and phosphatase inhibitors (Roche). Lysates were incubated for 15-minutes on ice, clarified, combined and split into equal amounts for parallel immunoprecipitations, each with 2ug neurofibromin-D antibody (Santa Cruz), rotating overnight at 4°C. Immunoprecipitations were immobilized on Protein A beads, rotating 1 h at 4°C, then washed 3x with IP buffer followed by 3x with ubiquitination wash buffer (50mM Tris-HCl pH 7.5, 5mM MgCl2). To assess NF1 ubiquitination catalyzed by Cul3 directly, exogenous HA-tagged Cul3 was expressed in NIH3T3 cells. Cells were stimulated with serum for 2 minutes and Cul3 was in ice-cold IP buffer for 2 hours and immobilized on protein A beads, washed 3x with IP buffer and washed 3x with ubiquitination wash buffer (50mM Tris-HCl pH 7.5, 5mM MgCl2) prior to in vitro ubiquitination reactions. Ubiquitination reactions were assembled containing the following and mixed with NF1-IP-beads: 50mM Tris-HCl pH.7.5, 5mM MgCl2, 2mM ATP (Sigma), 10mM phosphocreatine (Sigma), 3.5 U/ml creatine phosphokinase (Sigma), 1uM Ubiquitin aldehyde (Boston Biochem), 5ug ubiquitin (Boston Biochem), 220ng human E1 (Boston Biochem), 500ng E2 (UbcH5a) (Boston Biochem), and HA-Cul3 IP or control beads from an IgG immunoprecipitation as shown. Reactions were incubated at 37°C for 90 minutes and stopped by boiling in Laemmli buffer for 5 minutes. Western blots were immunoblotted with neurofibromin antibody (Bethyl). For ubiquitination reactions using Nrf2 as a substrate, GFP-Nrf2 was expressed in 293T cells and immunopurifed with a GFP-antibody (Invitrogen). Ubiquitination reactions were set up as described above.
Generation of constructs
DN-CUL3 (amino acids 1–418) and DN-CUL4A (amino acids 1–439) were cloned by PCR amplification including an N-terminal HA epitope. All fragments were cloned into pENTR-D TOPO (Invitrogen) according to the manufacturer’s instructions to generate pENTR-D clones containing HA-tagged DN-cullins. Mutant DN-Cul3 (L52AE55A) was generated using a site-directed mutagenesis kit (Clontech). Entry clones were subcloned into pLenti TO-puro destination vector (49) via Gateway-mediated LR recombination (Invitrogen) to generate lentiviral expression vectors. Clones expressing full-length cDNAs of KBTBD7, KLHL21, LacZ and Nrf2 in the pDONR223 vector were obtained from the Human Orfeome collection (Dana Farber Cancer Center) and subcloned into the lentiviral expression vector pHAGE-C-HA-FLAG (W. Harper, Harvard Medical School) by Gateway recombination using LR-clonase II (Invitrogen) according to the manufacturer’s instructions. Nrf2 was cloned into pHAGE N-GFP-blast expression vector (W. Harper). All final sequences were verified.
Retroviral and lentiviral infections
The lentiviral vector pLKO containing the following shRNAs were used: shCul3 (CCGGCGTGTTATAGACCACTC), shCul1 (CCGCACTTAAATCAATACATT), shCul2 (GCATCCAAG-TTCATATACTAA), shCul4B (GCTGTCTGATTTGCAAATTTA), shCul5 (CGAGAGTCCTATGTTAATCTT), shKBTBD7 (GAATTAAAGGACAGGCCAT). The retroviral vector pSUPER-Retro containing the following shRNAs were used: Cul3#1 (CCTCCAAGAATCCTTCAATAA), Cul3#2 (GCGAATAATGAAGTCTAGGAA) Cul3#3 (CGAGATCAAG-TTGTACGGTAT); Cul3#4 (CCAGGGCTTATTGGATCTAAA). Lentiviruses were produced by transfection of the 293T packaging cell line with the appropriate lentiviral expression vector, pLKO for shRNAs (TRC, Broad Institute) and pLentiTO or pHAGE-C-HA-FLAG for expression constructs, along with Δ8.2 (encoding gag, pol, and rev) and VSVG using Fugene 6 (Roche). Retroviruses were produced by transfection of the 293T packaging cell line with the appropriate retroviral expression vector and a vector encoding a replication-defective helper virus, pCL-Eco (Imgenex), using Fugene 6 (Roche). Virus-containing media was used to transduce target cells. 2 ug/ml puromycin was used to select stably transduced cell lines.
Identification of neurofibromin-interacting BTB proteins
Briefly, 100 15-cm plates of serum-deprived T98G GBM cells were pretreated with proteasome inhibitors for 2 hrs, and restimulated fetal calf serum. Cells were rinsed 3x with PBS and lysed in ice-cold 0.3% CHAPs buffer (0.3% CHAPS, 10mM HEPES pH7.5, 120nM NaCl, 1mM EDTA, 10mM sodium pyrophosphate, 50mM sodium fluoride) supplemented with protease and phosphatase inhibitors (Roche). Lysates were incubated for 15-minutes on ice, clarified, pre-cleared 2x with agarose beads, 2x with protein A beads for 1 h each time, and split into equal amounts for parallel immunoprecipitations with 50ug of neurofibromin antibodies (sc-67 or sc-68; Santa Cruz), rotating overnight at 4°C. Immunoprecipitations were immobilized on 150ul Protein A beads, rotating 2h at 4°C, and washed 5x with IP buffer. Samples were separated by SDS-PAGE on 4–12% gradient mini gels, and proteins were visualized with Coomassie Safely Blue stain (Invitrogen). The experimental lines were excised into small fragments, rinsed in 10% acetonitrile and submitted for mass spectrometry analysis at the BIDMC Mass Spectrometry Core facility.
Quantitative PCR
To detect shRNA-mediated knockdown of Kbtbd7 and KBTBD7 in mouse and human cell experiments, respectively, total RNA was extracted from cells using Trizol (Invitrogen), and cDNA was synthesized using qScript cDNA Synthesis Kit (Quanta), following the manufacturer’s instructions. Quantitative PCR was performed using PerfeCTa Sybr Green Mix (Quanta) following the manufacturer’s instructions. Expression levels of Kbtbd7 were normalized against Gapdh expression, and KBTBD7 expression was normalized to HPRT1 expression. The following primers were used: Kbtbd7_F (5’-aagagcactacctggga-3’), Kbtbd7_R (5’-cttggcaaccactgacttgt-3’), Gapdh_F (5’-actccactcacggcaaattc-3’), Gapdh_R (5-tctccatggtggtgaagaca-3’), KBTBD7_F (5’-cagtccgctcactctctatgt-3’), KBTBD7_R (5’-agacgccttcgaccatcac-3’), HPRT1_F (5’-gccggctccgttatgg-3’), HPRT1_R (5’-aacctggttcatcatcactaatca-3’).
Ras activation analysis
Primary MEFs were serum-starved by culturing to confluency for 4 days in DMEM supplemented with 10% FCS, then changing media to 0.1% FCS for 3 additional days. Cells were then resuspended, washed, and plated in serum-free DMEM overnight. Cells were stimulated with 6uM LPA for the times indicated. Ras-GTP levels were detected using a Ras activation assay, following the manufacturer’s instructions (Upstate). Ras activation was quantified by using DNA j software to assess the amount of Ras that was pulled down by GST-Ras-binding domain fusion protein, divided by total levels of Ras in the lysates. Values at 5 and 15 minutes were then divided by values observed in unstimulated control cells to establish relative levels of Ras activation.
Proliferation Studies
Quiescent MEFs were split to a density of 150,000 cells/well of a six-well dish in triplicate, and then stimulated with serum. At each indicated time, cells were trypsinized and counted. Fold increase in cell number ratios were calculated by dividing the average cell number of three plates in serum at indicated time points by the average cell number of three plates in serum-free medium after 18hrs.
Colony Growth in Soft Agar
Soft agar assays were performed as previously described(50).
Xenograft Tumor Studies
All animal procedures were approved by the Center for Animal and Comparative Medicine at the Harvard Medical School in accordance with the NIH Guild for the Care and Use of Laboratory Animals and the Animal Welfare Act. Subcutaneous implantations were carried out in 6-week old athymic female nude mice (Charles River) by injecting 106 cells in 100ul of PBS (6 or 8 injections per condition as described in the text). Two weeks after injection (day 0), the width and length of the tumor were measured twice weekly by caliper, and the volume was calculated with the following formula: volume = (length × width2)π/6. Tumors were monitored for 5 months.
Statistical Analyses
If data were normally distributed, statistical analysis was done by Student’s t-test. Otherwise we used the Mann-Whitney U test. Specific tests are noted in the text and figure legends. All numerical data, including error bars represent the mean ± standard deviation (SD) of experiments done in triplicate.
Supplementary Material
SIGNIFICANCE.
This study identifies the ubiquitin ligase complex that controls the regulated and pathogenic destruction of the NF1 tumor suppressor protein. These observations provide a molecular framework for developing potential therapies for glioblastoma, where neurofibromin is chronically destabilized and reveal a new strategy to attenuate Ras, which has broader therapeutic implications.
Acknowledgements
We thank Wade Harper and William Hahn for helpful discussions. We thank Andrew Wilkins for pSUPER-RETRO-based Cul3 shRNA constructs directed against Cul3, as well as pcDNA constructs containing full-length cullin cDNAs used for further sub-cloning. This work was supported bythe Department of Defense (W81XWH-08-1-0136) and the NCI (R01 CA111754).
Footnotes
Disclosure of Potential Conflicts of Interest: No conflicts of interest were disclosed.
References
- 1.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 3.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer [Internet] Nature Publishing Group. 2011;11:761–774. doi: 10.1038/nrc3106. Available from: http://www.nature.com/nrc/journal/v11/n11/full/nrc3106.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 5.Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, et al. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- 6.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med. 2010;16:286–294. doi: 10.1038/nm.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- 8.Riccardi VM. Neurofibromatosis: Phenotype, Natural History, and Pathogenesis, Second Edition. Second. Baltimore and London: The Johns Hopkins University Press; 1992. [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The I, Murthy AE, Hannigan GE, Jacoby LB, Menon AG, Gusella JF, et al. Neurofibromatosis type 1 gene mutations in neuroblastoma. Nat Genet. 1993;3:62–66. doi: 10.1038/ng0193-62. [DOI] [PubMed] [Google Scholar]
- 14.Hölzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142:218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messaien L, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittaker SR, Theurillat J-P, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andersen LB, Fountain JW, Gutmann DH, Tarlé SA, Glover TW, Dracopoli NC, et al. Mutations in the neurofibromatosis 1 gene in sporadic malignant melanoma cell lines. Nat Genet. 1993;3:118–121. doi: 10.1038/ng0293-118. [DOI] [PubMed] [Google Scholar]
- 18.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 19.Sherr CJ, Weber JD. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 20.Bloom J, Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin Cancer Biol. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nature Publishing Group. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 23.Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev. 2003;17:449–454. doi: 10.1101/gad.1054703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 25.Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim Biophys Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 26.Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- 27.Furukawa M, He YJ, Borchers C, Xiong Y. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol. 2003;5:1001–1007. doi: 10.1038/ncb1056. [DOI] [PubMed] [Google Scholar]
- 28.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 30.Lowy DR, Willumsen BM. Function and regulation of ras. Annu. Rev. Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 31.McEvoy JD, Kossatz U, Malek N, Singer JD. Constitutive turnover of cyclin E by Cul3 maintains quiescence. Mol Cell Biol. 2007;27:3651–3666. doi: 10.1128/MCB.00720-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu L, Wei Y, Reboul J, Vaglio P, Shin T-H, Vidal M, et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–321. doi: 10.1038/nature01985. [DOI] [PubMed] [Google Scholar]
- 33.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. PNAS. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pintard L, Willis JH, Willems A, Johnson J-LF, Srayko M, Kurz T, et al. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature. 2003;425:311–316. doi: 10.1038/nature01959. [DOI] [PubMed] [Google Scholar]
- 35.Geyer R, Wee S, Anderson S, Yates J, Wolf DA. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell. 2003;12:783–790. doi: 10.1016/s1097-2765(03)00341-1. [DOI] [PubMed] [Google Scholar]
- 36.Stogios PJ, Downs GS, Jauhal JJS, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, et al. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–594. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 38.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 39.Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, MacCoss MJ, et al. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat Cell Biol. 2006;8:348–357. doi: 10.1038/ncb1381. [DOI] [PubMed] [Google Scholar]
- 40.Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, et al. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 42.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 43.Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, et al. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, et al. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 48.Brugarolas J, Bronson RT, Jacks T. p21 is a critical CDK2 regulator essential for proliferation control in Rb-deficient cells. J Cell Biol. 1998;141:503–514. doi: 10.1083/jcb.141.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS ONE. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci USA. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.