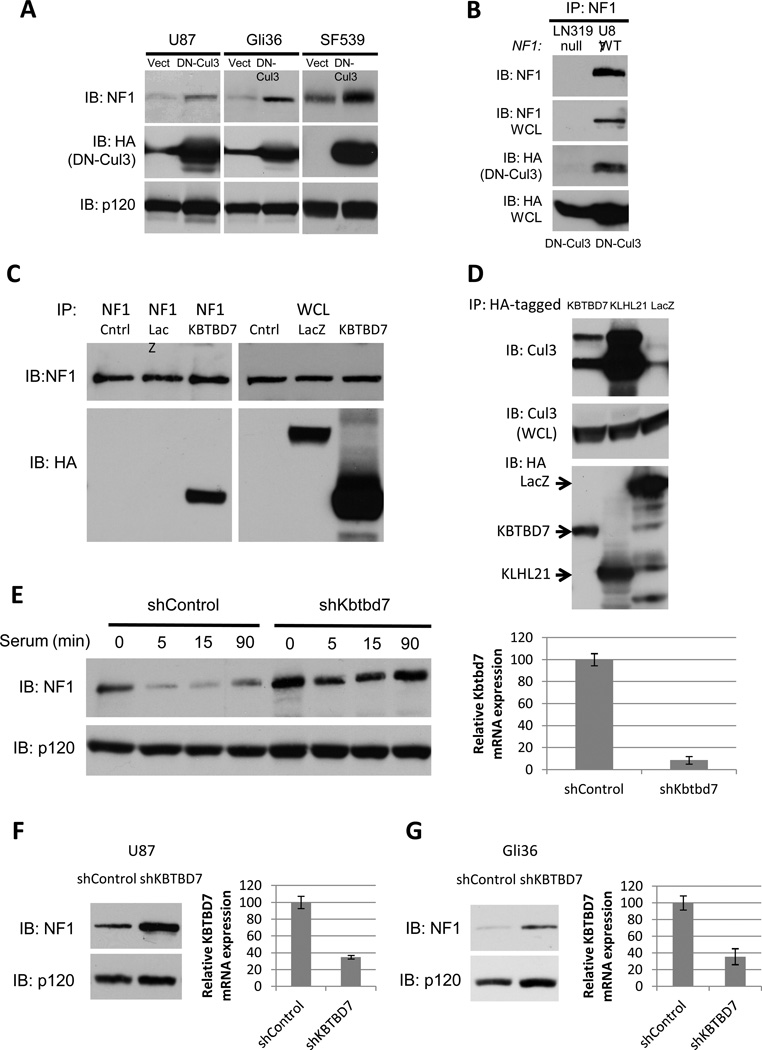
Figure 4. Cul3 and the adaptor protein KBTBD7 destabilize neurofibromin in GBM cells.
A) Immunoblot of neurofibromin, RasGAPp120 and HA-tagged dominant-negative Cul3 (DN-Cul3) from GBM cell lines expressing lentiviral HA-DN-Cul3 or a vector control.
B) Immunoblot of neurofibromin IP from NF1-wild type (U87) or NF1-null (LN319) GBM cells expressing HA-DN-Cul3. HA blot indicates that HA-DN-Cul3 associates with neurofibromin in U87 cells.
C) Immunoblot of neurofibromin IP from proteasome-inhibited, serum-stimulated cells expressing HA-KBTBD7 or HA-LacZ. HA blot indicates that KBTBD7 associates specifically with neurofibromin.
D) Immunoblot of endogenous Cul3 co-precipitating with HA-tagged BTB proteins KBTBD7 and KLHL21 expressed in 293T cells. Total Cul3 levels are shown as a loading control.
E) Left, immunoblot of neurofibromin degradation in serum-starved NIH3T3 fibroblasts stimulated with 10% serum for the times shown in the absence or presence of a short hairpin RNA specific for Kbtbd7. RasGAPp120 is shown as a loading control. Right, relative expression of Kbtbd7 mRNA as assessed by normalized quantitative PCR.
F) Left, immunoblot of neurofibromin in U87 GBM cells expressing a control or KBTBD7-specific shRNAs. RasGAPp120 is shown as a loading control. Right, relative expression of KBTBD7 mRNA as assessed by normalized quantitative PCR.
G) Left, immunoblot of neurofibromin in control Gli36 GBM cells or cells expressing a KBTBD7-specific shRNA. RasGAPp120 is shown as a loading control. Right, relative expression of KBTBD7 mRNA as assessed by normalized quantitative PCR.