Abstract
Objective
The aim of this study was to evaluate, by PCR-RFLP and Real-time PCR, the yield and quality of genomic DNA collected from buccal cells by mouthwash after different storage times at room temperature.
Material and Methods
A group of volunteers was recruited to collect buccal cells using a mouthwash solution. The collected solution was divided into 3 tubes, one tube were used for immediate extraction and the remaining received ethanol and were kept at room temperature for 4 and 8 days followed by DNA extraction. The concentration, purity and integrity of the DNA were determined using spectrophotometry and electrophoresis. DNA quality differences among the three incubation times were also evaluated for genotyping EGF +61 A/G (rs 4444903) polymorphism by PCR-RFLP and for IRF6 polymorphism (rs 17015215) using Real-Time PCR.
Results
There was no significant difference of DNA yield (p=0.75) and purity (p=0.86) among the three different incubation times. DNA obtained from different incubation times presented high-molecular weight. The PCR-RFLP and Real time PCR reactions were successfully performed for all DNA samples, even those extracted after 8 days of incubation. All samples genotyped by Real-Time PCR presented C allele for IRF6 gene polymorphism (homozygous: CC; heterozygous: CT) and the C allele was used as a reference for Ct values. The samples presented the same genotype for the different times in both techniques.
Conclusion
We demonstrated that the method described herein is simple and low cost, and that DNA can be extracted and PCR amplified after storage in mouthwash solution at room temperature.
Keywords: DNA, Saliva, PCR, Genetic polymorphism
INTRODUCTION
In the last years, the advances in molecular genetics research provided substantial progress regarding the identification of genes involved in the pathogenesis of human alterations and diseases2. Molecular epidemiological studies regarding the association of mutation/polymorphisms with orofacial diseases, including oral clefts, developmental dental alterations and oral diseases (i.e. caries lesion and periodontal disease), advanced on many levels14,19,20.
In epidemiological studies10, most of them using polymerase chain reaction (PCR) technique17, obtaining high-quality genomic DNA is critical. In most cases the preferred source of material is peripheral blood. Blood sampling, however, may be problematic in cases such as extreme illness or elderly people, babies and people that are reluctant to be submitted to this invasive procedure. For this reason, buccal cell collection is gaining recognition as an alternative specimen source for DNA isolation in clinical and research testing6. Additionally, clinical validation studies have shown that DNA diagnostic results from patient-matched buccal cells and whole blood are comparable3.
Buccal cells can be obtained for DNA isolation using mouthwashes, cytobrushes, swabs, treated cards and whole saliva collection. Mulot, et al.12 (2005) compared cytology brushes, mouthwash and treated cards for DNA collection and concluded that cytobrush appears to be the most appropriate method with good quality and high security in multicenter studies. On the other hand, another study comparing oral rinse, cytobrush and swab using OrageneTM DNA collection kit to extract DNA showed that oral rinse provided sufficient DNA quantity and quality better than the buccal swab and brush techniques16.
Commercially available DNA isolation kits are produced mainly in industrialized countries, and may not be readily available at affordable prices in developing and underdeveloped countries1. In order to establish DNA collection of a large number of samples for epidemiological studies, we used a simple and cost-efficient protocol1,7. The aim of this study was to evaluate, by restriction fragment length polymorphism-PCR (PCR-RFLP) and Real-time PCR, the yield and quality of genomic DNA collected from buccal cells by mouthwash after different storage times at room temperature.
MATERIAL AND METHODS
A group of 12 volunteers of both genders (with ages ranging from 20 to 54 years) were enrolled to collect buccal cells using a mouthwash solution. Ethical approval was obtained from the Human Ethics Committee of the Health Department of the city of Rio de Janeiro, Rio de Janeiro, Brazil (113/09). Informed consent was obtained from all participating individuals. The individuals were instructed to brush their teeth or rinse the mouth with water for at least 2 h prior to saliva collection.
Saliva collection
Buccal cells were collected by rinsing the mouth for 60 s with 15 mL of saline and expectorating the rinse in a 50 mL propylene tube. In order to assess the DNA integrity over time, the mouthwash solution was divided into 3 tubes.
One tube was used for immediate extraction (T0) and the two remaining received ethanol up to a 70% final concentration and were kept at room temperature for 4 (T4) or 8 (T8) days followed by DNA isolation.
DNA extraction
Genomic DNA was extracted from buccal epithelial cells following a modified protocol reported by Aidar and Line1 (2007). Briefly, after incubation the tubes were centrifuged for 10 min at 550 xg at room temperature to pellet the bucal cells. The supernatant was discarded and 1 mL of extraction solution (Tris-HCl 10 mM, pH 7.8; EDTA 5 mM; SDS 0.5 %) containing proteinase K (100 ng/mL) (Invitrogen, Grand Island, NY, USA) was added to cause lysis of cells and proteins. After overnight incubation the non-digested proteins were removed by adding 400 µL of 10 M ammonium acetate. The solutions were mixed by gently reversing the tube 20 times and centrifuged at 12,000 xg for 15 min. Supernatant was separated into two clean 1.5 mL microtubes (700 µL) and DNA was precipitated with 700 µL of isopropanol at -20ºC for 30 minutes. After centrifugation for 20 min at 12,000 xg at 4ºC, the supernatant was poured off and the pellet washed with 1 mL of 70 % ethanol. Ethanol was decanted carefully after centrifugation at 12,000 xg for 15 min at 4ºC, the tubes were reversed and allowed to air-dry during 45 to 60 min on an absorbent paper. DNA was resuspended in 100 µL of TE buffer (10 mM Tris (pH 7.8) and 1 mM EDTA) and stored at -20ºC.
Criteria for evaluating DNA extraction after different incubation times
1. Concentration measurement assays
The concentration and purity of the DNA was determined by spectrophotometry using NanoDrop 2000c. DNA concentration was evaluated at 260 nm and the ratio of readings at 260 nm and 280 nm was used to estimate the DNA purity.
2. DNA integrity
The integrity of genomic DNA for 0, 4- and 8-day incubation times was assessed by resolving DNA on a 0.8 % agarose gel by electrophoresis, followed by visualization with ethidium bromide staining.
3- DNA quality and polymorphism genotyping.
PCR Assays and RFLP
In order to evaluate DNA quality for polymorphism genotyping by PCR-RFLP DNA samples of each incubation time were assayed as previously described by Shahbazi18 (2002), for the polymorphism in the EGF (+61 A/G) gene. The PCR reaction was carried out in a 30 µL mixture containing 160 ng of genomic DNA, 1x Dream Taq Buffer (Fermentas, Maryland, NY, USA), 0.2 mM of dNTPs (Fermentas), 0.3 µM of each primer (Forward: 5' TGTCACTAAAGGAAAGGAGG3'; Reverse: 5' TTCACAGAGTTTAACAGCCC 3' - Invitrogen, Grand Island, NY, USA) and 1 U Dream Taq DNA Polymerase (Fermentas, Maryland, NY, USA). The cycling conditions comprised an initial denaturation step for 10 min at 96ºC, followed by 44 cycles consisting of three steps: 96ºC for 30 sec, 61ºC for 30 sec and 72ºC for 1 min. A final extension step was performed at 72ºC for 10 min.
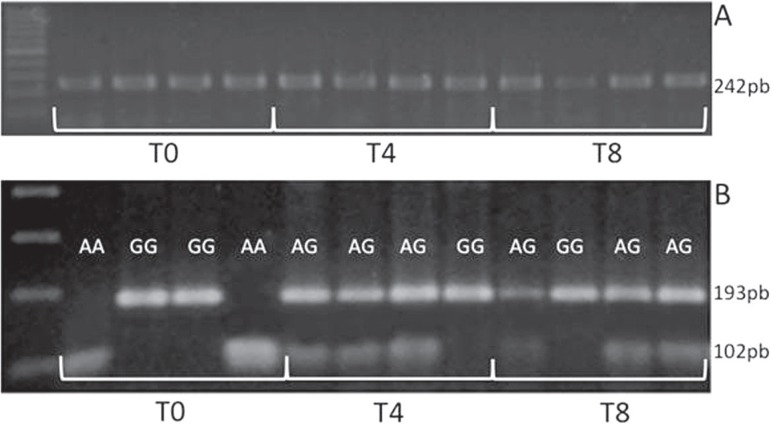
The amplified fragment (242 bp) was digested with 2 U of Alu I (New England Biolabs, Beverly, MA, USA) at 37ºC overnight, electrophoresed on a 2.5% agarose gel stained with ethidium bromide and photographed under UV illumination. Four fragments (15, 34, 91 and 102 bp) were produced in the presence of A allele and three fragments (15, 34 and 193 bp) were produced in the presence of G allele. In the gel, the 15 and 39 bp fragments were not visible. Therefore, the G allele was differentiated from the A allele by visualization of a single 193 bp fragment instead of the 91 and 102 bp fragments.
TaqMan amplification
DNA quality differences among the three incubation times were also evaluated by Real-Time PCR. The cycle threshold (Ct) is defined as the number of cycles required for the fluorescent signal to cross the threshold. In a real time PCR assay a positive reaction is detected by accumulation of a fluorescent signal. Ct levels are inversely proportional to the amount of target nucleic acid in the sample.
Quantified DNA was diluted to 4 ng/mL and 6 ng of DNA were used to perform the genotyping of IRF6 polymorphism (C/T transition, rs 17015215) using Taqman probes15 in an Sequence Detection System instrument (Mx3005P system; Stratagene). Assays and reagents were supplied by Applied Biosystems (Foster City, CA, USA). Determination of amplification was computed as a Ct value using the MxPro-Mx3005p software supplied by the manufacturer.
Statistical analysis
Comparisons of the three incubation times were made to determine differences in mean total DNA yield and PCR and Real-Time amplification Ct under different sample storage times were tested using ANOVA.
RESULTS
The total DNA yield measured at 260 nm was 19.4±8.4 µg (11.1 - 38.8 µg), 24.2±13.9 µg (9.0 - 51.5 µg) and 21.9±13.8 µg (9.3 - 46.4 µg) and the mean OD 260/280 ratio was 2.03±0.04 (1.97-2.12), 2.01±0.05 (1.94-2.05) and 2.02±0.09 (1.87-2.15) for T0, T4 and T8 respectively.
This results indicated that there was no significant difference between the three different incubation times for DNA yield (p=0.75) and purity (p=0.86).
Gel electrophoresis was used to visualize the presence of high-molecular weight DNA. DNA obtained from different incubation times presented high-molecular weight for all three times (Figure 1).
Figure 1.
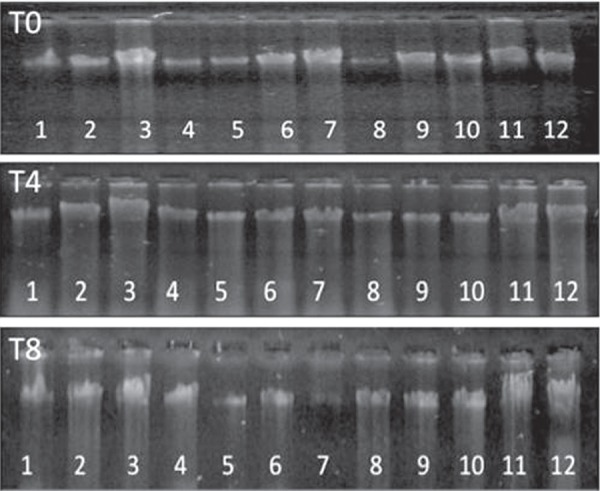
Genomic DNA electrophoresis from DNA obtained from different incubation times (T0, T4 and T8)
The PCR-RFLP-reactions were successfully performed for all DNA samples, even using DNA samples extracted after 8-day incubation time. Four different samples representing PCR products and genotyping by restriction enzymes are illustrated in Figure 2.
Figure 2.
Epidermal growth factor (EGF) polymorphism (rs 4444903) genotyped by polymerase chain reaction - restriction fragment length polymorphism (PCR-RFLP) using DNA extracted from different incubation times (T0, T4 and T8); A) PCR amplicons and B) digestion with Alu I
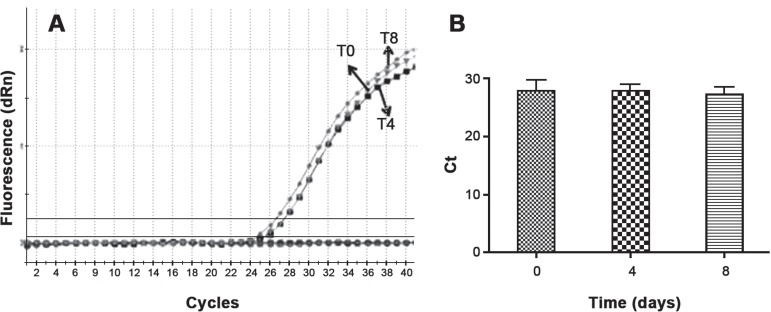
All samples were also successfully genotyped by Real-Time PCR using TaqMan probes. All of them presented C allele for IRF6 gene polymorphism (homozygous - CC and heterozygous - CT) and the C allele was used as a reference for Ct values. The Ct values ranged from 26 to 28 and the Ct mean values are presented on Figure 3B for T0, T4 and T8 incubation times. There is no difference between mean Ct values and incubation time (p=0.73) (Figure 3B).
Figure 3.
Genotyping of IRF6 (rs17015215) polymorphism by real time polymerase chain reaction (PCR). A) amplification plot of the same DNA in different time incubation and B) mean Ct values in different time incubation
The samples presented the same genotype after the different times for both techniques.
DISCUSSION
Exfoliated buccal epithelial cells in saliva are a very promising alternative source of DNA because they can be obtained using self-administered, noninvasive, and relatively inexpensive techniques5,10,13. Collection of samples for DNA extraction is a critical procedure as it is time-consuming and may involve ethical aspects. It is, therefore, desirable that this procedure becomes more simple and low-cost1. The protocol proposed here has some advantages, for being simple, low cost and not using phenol-chloroform as in other techniques11,16,21. Our results showed that the buccal cells provided satisfactory amount of DNA. Our mouthwash method presented similar values from other mouthwash methods6,8. In our sample, the amount of extracted DNA had inter-individual variation depending on the buccal mucosa.
Previous studies reported the greatest similar efficiency values for mouthwash and cytobrush, based on the same DNA extraction method4,8. However, saline mouthwash has lower cost when compared with cytobrush.
DNA quality can be affected by the collection method (primary integrity and protein contamination) and by the isolation method (integrity and protein, organic, salt and alcohol contamination)16. In our study, the mean OD 260/280 ratio was 2.02, indicating that in most cases the bulk of proteins were successfully removed by ammonium acetate precipitation.
The storage time at room temperature had few effects on DNA stability at room temperature. This could be crucial for large-scale epidemiological studies, which require a time for collection logistics. On the other hand, the main purpose of collecting DNA is for future studies and our RFLP-PCR and Real-Time PCR was successfully performed for the three incubation times.
CONCLUSION
Our results demonstrated that the method described here is low-cost and simple to be performed by dental and medical researchers. DNA can be extracted and PCR amplified after storage in mouthwash solution at room temperature. Several samples can be processed at the same time and the extracted DNA yields sufficient material for many genetic studies.
Acknowledgments
ACKNOWLEDGMENTS
This study was supported in part by grants from the Nacional Council for Scientific and Technological Development (CNPq grant 482457/2009-0) and the Rio de Janeiro Research Foundation (FAPERJ grant E-26/102.729/2008).
REFERENCES
- 1.Aidar M, Line SR. A simple and cost-effective protocol for DNA isolation from buccal epithelial cells. Braz Dent J. 2007;18:148–152. doi: 10.1590/s0103-64402007000200012. [DOI] [PubMed] [Google Scholar]
- 2.Bailleul-Forestier I, Molla M, Verloes A, Berdal A. The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur J Med Genet. 2008;51:273–291. doi: 10.1016/j.ejmg.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.De Vries HG, Collèe JM, van Veldhuizen MH, Achterhof L, Smit Sibinga CT, Scheffer H, et al. Validation of the determination of deltaF508 mutations of the cystic fibrosis gene in over 11,000 mouthwashes. Hum Genet. 1996;97:334–336. doi: 10.1007/BF02185766. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–696. [PubMed] [Google Scholar]
- 5.Harty LC, Shields PG, Winn DM, Caporaso NE, Hayes RB. Self-collection of oral epithelial cell DNA under instruction from epidemiologic interviewers. Am J Epidemiol. 2000;151:199–205. doi: 10.1093/oxfordjournals.aje.a010188. [DOI] [PubMed] [Google Scholar]
- 6.Heath EM, Morken NW, Campbell KA, Tkach D, Boyd EA, Strom DA. Use of buccal cells collected in mouthwash as a source of DNA for clinical testing. Arch Pathol Lab Med. 2001;125:127–133. doi: 10.5858/2001-125-0127-UOBCCI. [DOI] [PubMed] [Google Scholar]
- 7.Juárez-Cedillo T, Sánchez-García S, Mould-Quevedo JF, García- Peña C, Gallo JJ, Wagner FA, et al. Cost-effective analysis of genotyping using oral cells in the geriatric population. Genet Mol Res. 2010;9:1886–1895. doi: 10.4238/vol9-3gmr939. [DOI] [PubMed] [Google Scholar]
- 8.King IB, Satia-Abouta J, Thornquist MD, Bigler J, Patterson RE, Kristal AR, et al. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev. 2002;11:1130–1133. [PubMed] [Google Scholar]
- 9.Koni AC, Scott RA, Wang G, Bailey ME, Peplies J, Bammann K, et al. DNA yield and quality of saliva samples and suitability for large-scale epidemiological studies in children. Int J Obes (Lond) 2011;35:S113–S118. doi: 10.1038/ijo.2011.43. [DOI] [PubMed] [Google Scholar]
- 10.Le Marchand L, Lum-Jones A, Saltzman B, Visaya V, Nomura AM, Kolonel LN. Feasibility of collecting buccal cell DNA by mail in a cohort study. Cancer Epidemiol Biomarkers Prev. 2001;10:701–703. [PubMed] [Google Scholar]
- 11.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–724. [PubMed] [Google Scholar]
- 12.Mulot C, Stücker I, Clavel J, Beaune P, Loriot MA. Collection of human genomic DNA from buccal cells for genetics studies: comparison between cytobrush, mouthwash, and treated card. J Biomed Biotechnol. 2005;2005:291–296. doi: 10.1155/JBB.2005.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nedel F, André DA, Oliveira IO, Tarquinio SB, Demarco FF. Buccal cells submitted to three different storage conditions before DNA extraction. J Appl Oral Sci. 2009;17:113–115. doi: 10.1590/S1678-77572009000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paranaiba LM, Bufalino A, Martelli-Junior H, Barros LM, Graner E, Coletta RD. Lack of association between IRF6 polymorphisms (rs2235371 and rs642961) and non-syndromic cleft lip and/or palate in a Brazilian population. Oral Dis. 2010;16:193–197. doi: 10.1111/j.1601-0825.2009.01627.x. [DOI] [PubMed] [Google Scholar]
- 15.Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, et al. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers NL, Cole SA, Lan HC, Crossa A, Demerath EW. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;19:319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos CF, Sakai VT, Machado MA, Schippers DN, Greene AS. Reverse transcription and polymerase chain reaction: principles and applications in dentistry. J Appl Oral Sci. 2004;12:1–11. doi: 10.1590/s1678-77572004000100002. [DOI] [PubMed] [Google Scholar]
- 18.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 19.Tannure PN, Kuchler EC, Lips A, Costa MC, Luiz RR, Granjeiro JM, et al. Genetic variation in MMP20 contributes to higher caries experience. J Dent. 2012;40:381–386. doi: 10.1016/j.jdent.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. A genome wide linkage scan for cleft lip and palate and dental anomalies. Am J Med Genet A. 2008;146A:1406–1413. doi: 10.1002/ajmg.a.32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh DJ, Corey AC, Cotton RW, Forman L, Herrin GL, Jr, Word CJ, et al. Isolation of deoxyribonucleic acid (DNA) from saliva and forensic science samples containing saliva. J Forensic Sci. 1992;37:387–395. [PubMed] [Google Scholar]