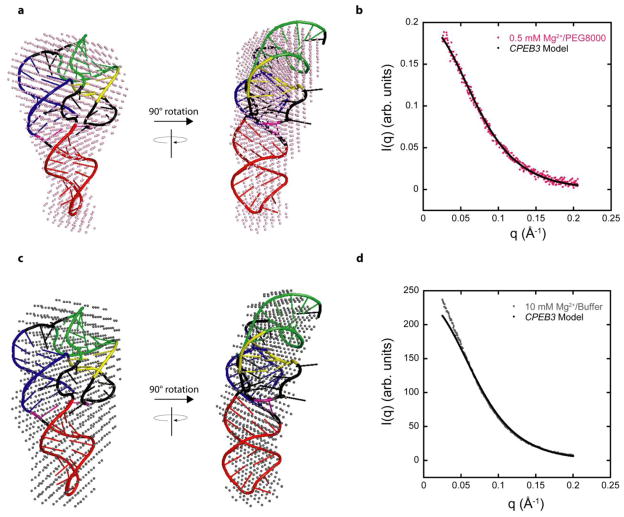
Figure 7. Model of CPEB3 ribozyme agrees well with SAXS data.
(a,c) SAXS reconstructions in (a) 0.5 mM Mg2+ and 20% PEG8000 (pink spheres) and (c) 10 mM Mg2+ and no additive (grey spheres) superimposed on the same CPEB3 ribozyme model. The CPEB3 ribozyme model was constructed from the HDV ribozyme crystal structure and color-coded according to the secondary structure in Figure 1. (b,d) Experimental scattering data for CPEB3 ribozyme in (b) 0.5 mM Mg2+ and 20% PEG8000 and (d) 10 mM Mg2+ and no additive plotted with the calculated scattering data from native state model. The calculated scattering profile for CPEB3 model was generated using FoXS server.36,37