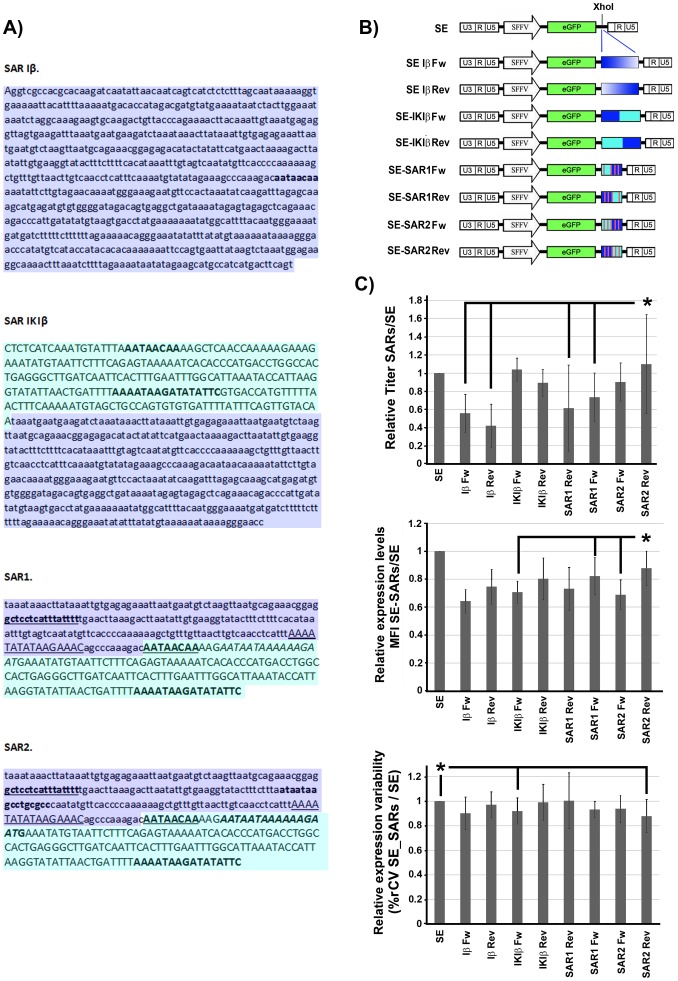
Figure 1. A 388/improved properties than the Interferon β SAR on K562 cells.
(A) Sequences of the different SARs elements used in the study. SAR Iβ contains 790 bp from the Interferon β locus (AL390882.12: nucleotides 84551–83762). SAR IgKIβ is a chimeric construct containing 240 bp from the Ig Kappa (IgK) consensus SAR sequence (capital case; NG_000834.1: nts 482307–482547) and 480 bp of the SAR Iβ (minor case; AL390882.12: nts 84357–83877). SAR1 and SAR2 synthetic elements are designed to contain 4 and 5 SARs recognition signatures (MRS) respectively. These MRS are embedded into 231 bp of the Iβ locus (AL390882.12; nts 84362–84131, purple) and 157 bp of the IgK locus (NG_000834.1; nts 482316-482473, green). The MRS as described by van Drunen et al [29] are: - 1 MRS from the Igk locus (in capital-bold), - 2 MRSs from the β-globin locus (β-globin overlapping: in capital-bold-italic and β-globin +11: in capital-underlined) and - 1 (SAR1) or 2 (SAR2) MRSs from the ψ-globin locus (ψ-globin overlapping: in bold-normal and the ψ-globin +144: in bold-underlined). (B) Schematic diagram of the different SE LVs harboring the different SARs. The SARs where inserted between the enhanced green fluorescence protein (eGFP) and the 3′ LTR. (C). Effects of the insertion of the different SARs elements on titer (top graph), expression levels (middle graph) and variability of expression (%rCV; bottom graph). All data are represented relative to the parental SE LV. Values represent mean of at least three separate experiments and the error bar indicates the standard error of the mean. (* = p<0.05; two tail unpaired Student t-test).