Fig. 2.
Cytology-based dihybrid screens for demonstrating VirE2–VirD4 complex formation.
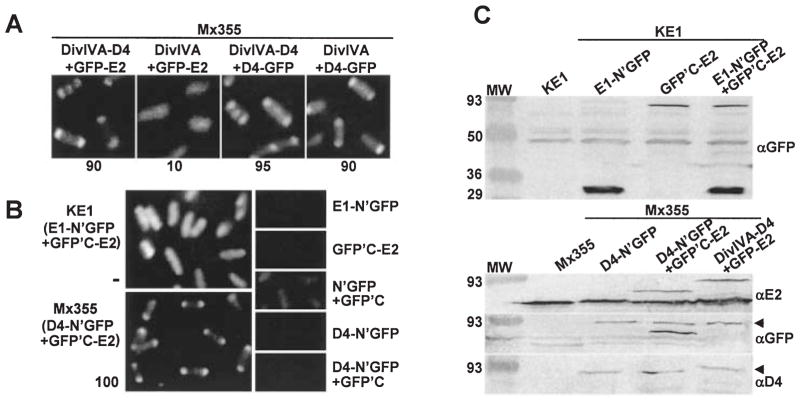
A. C2H: A348 (WT) cells producing the proteins indicated above each panel were examined 4 h post induction by fluorescence microscopy. The proteins indicated were synthesized from the following plasmids: DivIVA-D4 + GFP-E2 (pZD76, pZDB73); DivIVA + GFP-E2 (pKA76, pZDB73); DivIVA-D4 + D4-GFP (pZD76, pKA62); DivIVA + D4-GFP (pKA76, pKA62). The number below each panel represents the percentage of cells with polar fluorescence. Typically, ~ 15% of cells exhibiting DivIVA-dependent targeting in the C2H screen show fluorescence at the midcell.
B. BiFC: KE1 (ΔvirE) and Mx355 cells producing the proteins indicated adjacent to each panel were examined 4 and 10 h postinduction, respectively, by fluorescence microscopy. The proteins indicated were synthesized from the following plasmids introduced into the strains singly or in combination: E1-N’GFP (pZDB88); GFP’C-E2 (pZDB89); D4-N’GFP (pKAB64); N’GFP + GFP’C (pKVB35, pKVB39); D4-N’GFP (pKAB64); D4-N’GFP + GFPC’ (pKAB64, pKVB39).
C. Immunodetection of fusion proteins produced in KE1 (top panel) and Mx355 (bottom panel) at 10 h post induction. The proteins listed above each lane were synthesized from the following plasmids introduced into the strains singly or in combination: E1-N’GFP (pZDB88); GFP’C-E2 (pZDB89); D4-N’GFP (pKAB64); D4-N’GFP + GFPC’-E2 (pKAB64, pZDB89); and DivIVA-D4 + GFP-E2 (pKV42). The dark arrowheads point to distinct proteins that exhibit similar mobilities, e.g., VirD4-N’GFP (~ 92-kDa) and GFP-VirE2 (~ 90-kDa) (immunoreactive with anti-GFP antiserum), and VirD4-N’GFP and DivIVA-VirD4 (~ 93 kDa) (immunoreactive with anti-VirD4 antiserum).