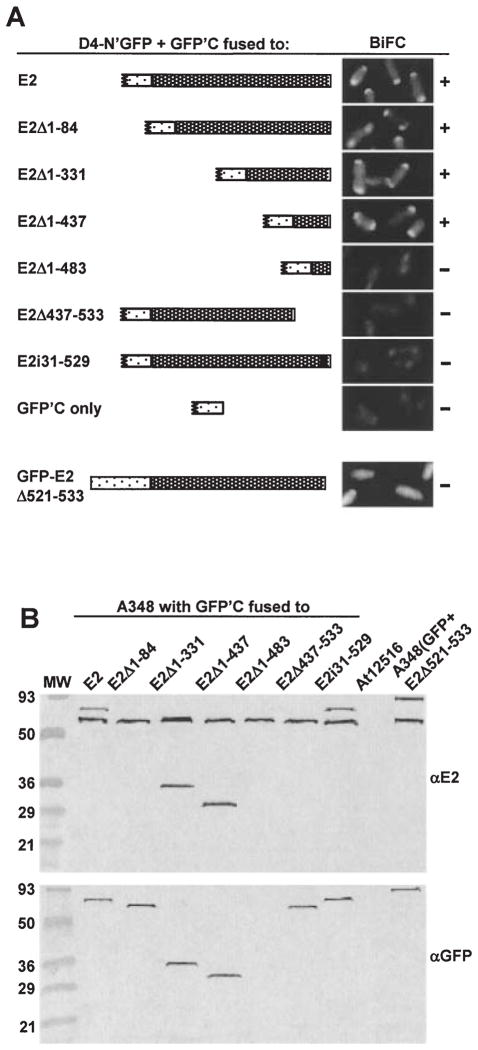
Fig. 4.
Localization of a VirD4 interaction domain to the C terminus of VirE2.
A. A348 producing VirD4-N’GFP and GFP’C fused to the VirE2 derivatives listed at the left and schematically represented were examined at 10 h (top two panels) or 18 h (bottom six panels) post induction by fluorescence microscopy. The proteins indicated were produced from the following plasmids: D4-N’GFP (pKAB64); GFPC’-E2 (pZDB89); GFPC’-E2Δ1–84 (pZDB838); GFPC’-E2Δ1–331 (pZDB834); GFPC’-E2Δ1–437 (pZDB836); GFPC’-E2Δ1–483 (pZDB122); GFPC’-E2Δ437–533 (pZDB846); GFPC’-E2i31–529 (pZDB827); GFPC’ (pKVB39); GFP-VirE2Δ524–533 (pZDB120). ‘ +’, fluorescent foci. ‘−’, no detectable foci, although these cells exhibited weak uniform fluorescence at t = 18.
B. Immunodetection of fusion proteins produced in A348 derivatives at 10 h postinduction. The proteins listed above each lane were synthesized from the IncP plasmids listed in (A). Blots were developed with the antisera listed at the right. The reactive species (~60-kDa) in all lanes, except At12516 (virE2−), reactive with the anti-VirE2 antisera is native VirE2 produced from pTi.