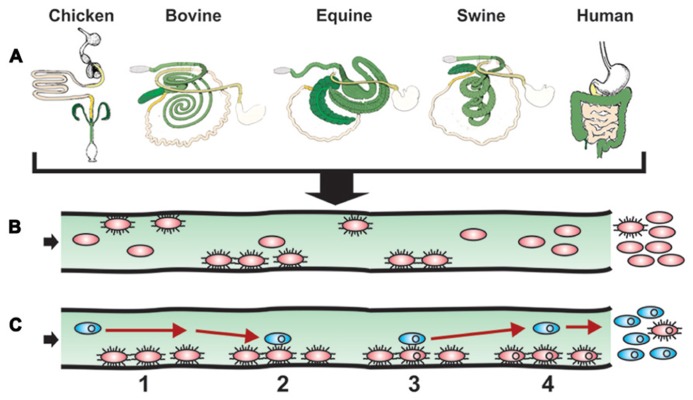
FIGURE 2.
Model of intestinal surface colonization and HGT. (A) The success story of Salmonella evolution and adaptation to a multitude of environments and hosts is illustrated by showing how diverse the intestinal tract anatomies of just a few warm blooded animals are when comparing chicken, bovine, equine swine, and human. In addition to the distinct anatomies, histological, immunological, and microbiota particularities further participate to create unique compartments in which varieties of Salmonella developed ways to live and multiply, with a simple escape route for future settlements. (B) S. enterica expressing specific allelic adhesins for the cognate host intestinal receptors and cellular targets initiate successful colonization. Other strains lacking such ligands will not be retained as well in the host. A resulting gastroenteritis and/or persistence will result in the multiplication and possible long-term excretion of such Salmonella strains (not shown). (C) Intestinal colonization mediated by specific allelic adhesins of Salmonella (red ovals) optimizes contact (event numbers and time span) with a constant flow of new bacteria (blue ovals), some carrying antimicrobial resistance genes on conjugative or mobilizable elements (small circles in ovals), resulting in increased HGT efficiency and antibiotic-resistant S. enterica colonizing carrier hosts. With time, excretion of antibiotic-resistant S. enterica will be excreted in greater numbers than antibiotic-susceptible S. enterica (not shown).