Figure 2.
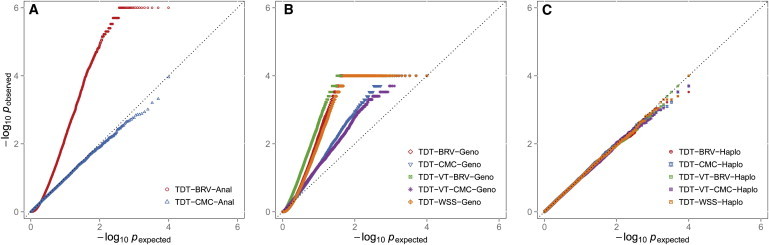
QQ Plot of Negative Natural Log p Values Obtained for Trio Data under the Null Hypothesis of No Association when the Variant Sites that Are Tested Are in Perfect LD
For each scenario, a total of 1,500 trios were analyzed and 20,000 replicates were generated. For the TDT-CMC and TDT-BRV, variants with MAF ≤ 1% were analyzed while for the TDT-VT-BRV, TDT-VT-CMC, and TDT-WSS, variants with MAF ≤ 5% were analyzed.
(A) Displays the results for the TDT-BRV and TDT-CMC when p values were obtained analytically (Anal).
(B) Displays the results for the TDT-BRV, TDT-CMC, TDT-VT-BRV, TDT-VT-CMC, and TDT-WSS. All p values were obtained empirically by performing 10,000 genotype (Geno) permutations for each replicate.
(C) Displays the results for the TDT-BRV, TDT-CMC, TDT-VT-BRV, TDT-VT-CMC, and TDT-WSS. All p values were obtained empirically by performing 10,000 haplotype (Haplo) permutations for each replicate.
