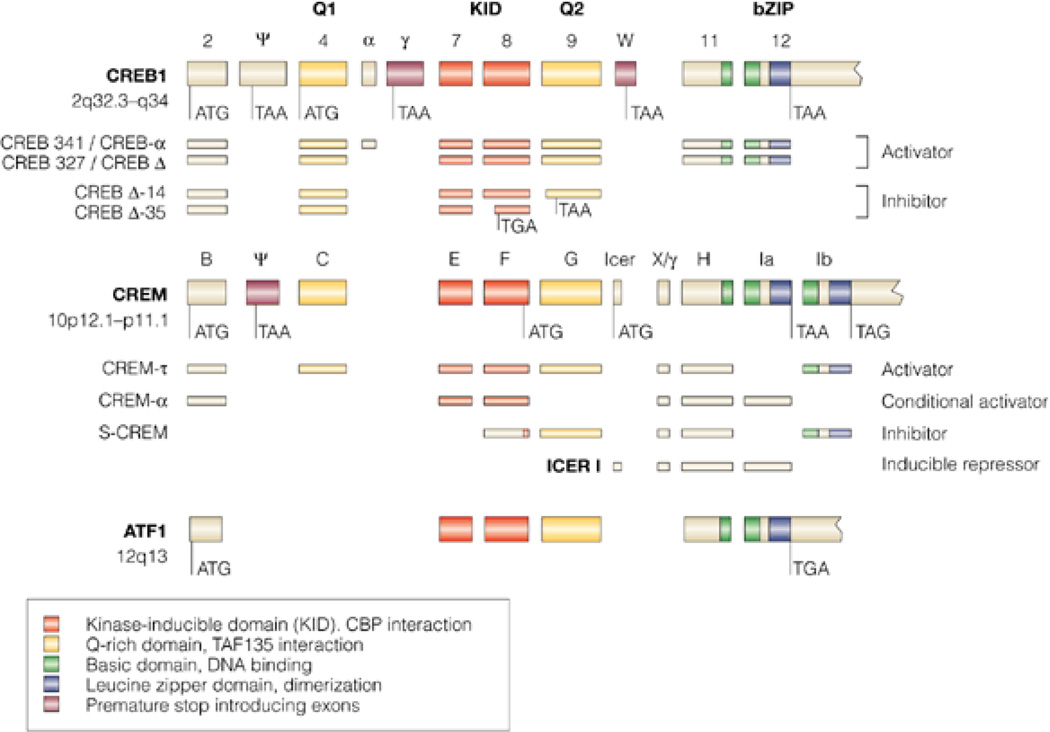
Figure 2. Genomic organization of the mammalian CREB family.
The consensus alignment of the genomic organization of human cyclic AMP response element (CRE)-binding protein (CREB), the cAMP response element modulator (CREM) and the activating transcription factor 1 (ATF1) was obtained by BLAST analysis of human GenBank sequences against the human genome and against each other. In cases for which human sequences were unavailable for published isoforms, the mouse or rat sequences were used. Homologous domains are in color (see key). In the CREB and CREM genes, several alternatively spliced exons exist, creating in-frame stop codons (TAA or TGA) leading to carboxy-terminally truncated proteins or giving rise to amino-terminally truncated proteins by using internal transcription-initiation sites downstream of the stop-introducing exon. A partial list of alternative splice products with divergent activating properties is shown. [From Mayr & Montminy (151)].