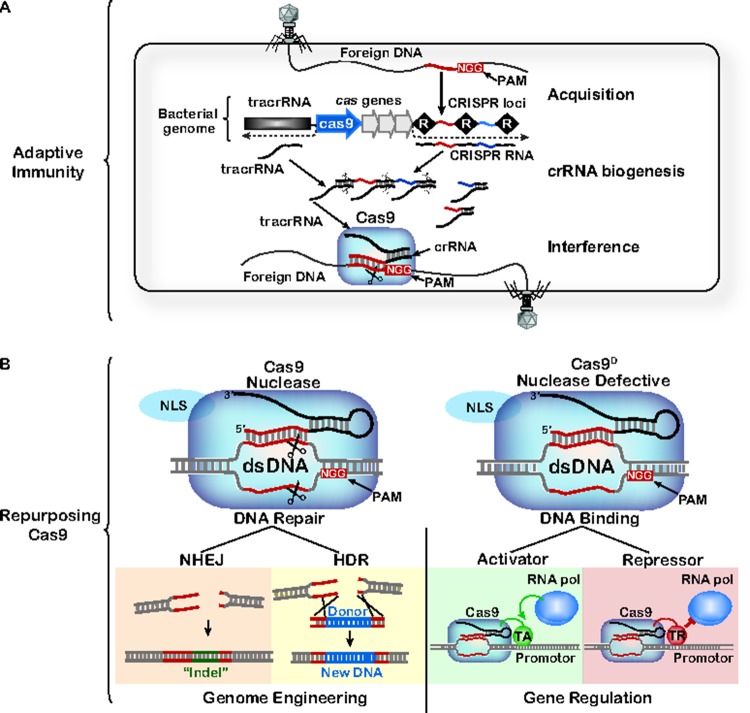
Figure 1. Repurposing RNA-guided nucleases from the CRISPR-mediated adaptive immune system in bacteria.
A) CRISPR-mediated adaptive immunity proceeds in three distinct stages: acquisition of foreign DNA, CRISPR RNA (crRNA) biogenesis, and target interference. Bacteria acquire resistance to viral and plasmid challengers by integrating short fragments of foreign nucleic acid (called protospacers) into CRISPR loci encoded in the bacterial genome. Protospacers are selected from regions of the genome that are flanked by a short sequence motif called a protospacer adjacent motif (PAM). CRISPR loci consist of a series of short repeats (R, black diamonds) and unique spacers (red and blue lines). CRISPR loci are transcribed and the RNA is processed into a library of small CRISPR-derived RNAs (crRNAs). In some CRISPR systems (i.e. Type II), a trans-activating CRISPR RNA (tracrRNA) is essential for RNA processing and for recognition by Cas9 (CRISPR-associated protein 9). Cas9 is an RNA-guided, dsDNA binding protein that uses two nuclease domains to cleave both strands of target DNA.
B) Cas9 targeting relies on PAM recognition and base pairing between the crRNA and the target DNA. The Cas9 nuclease can be easily programmed to target any DNA sequence with an adjacent PAM by designing a crRNA complementary to the target sequence. Genomic double-stranded DNA breaks are repaired by non-homologous end joining (NHEJ) or homology directed repair (HDR). NHEJ is error-prone, resulting in insertions or deletions (indels) that disrupt the target site. HDR relies on a donor template that can be used to deliver foreign DNA at a specific location. Alternatively, nuclease defective versions of Cas9 (Cas9D) have been tethered to transcription activators (TA) that promote gene transcription, or transcriptional repressors (TR) that inhibit transcription.