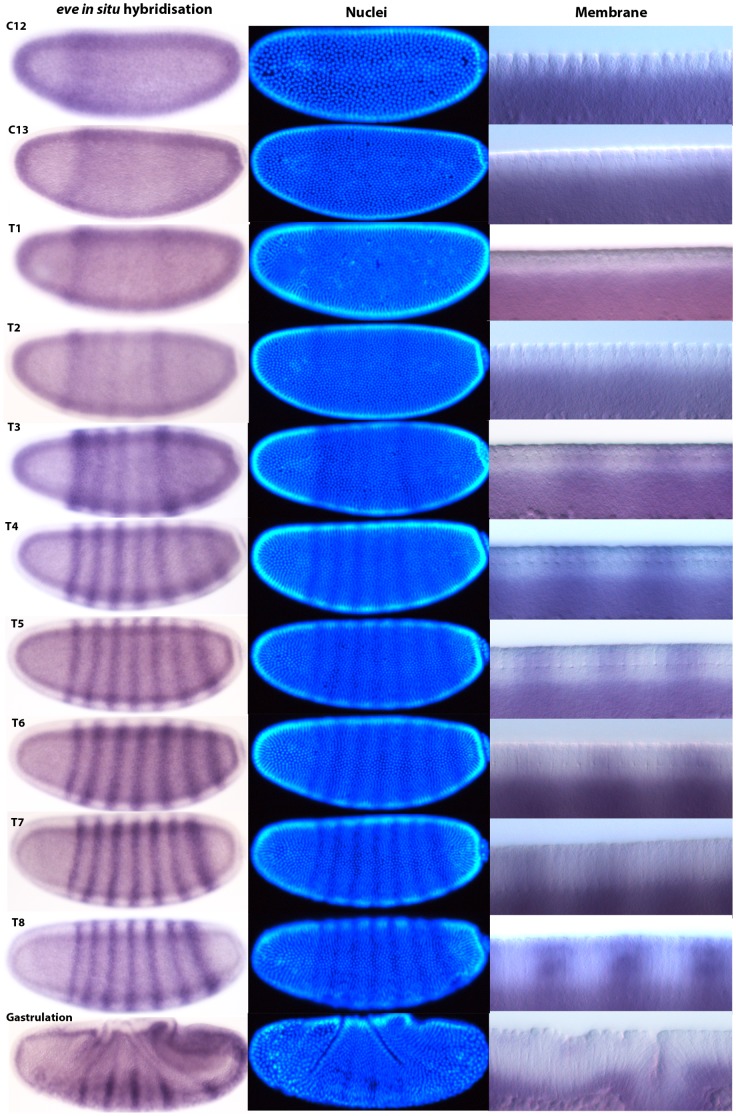
Figure 8. M. abdita eve mRNA expression staged using nuclei number, nuclear density, and membrane morphology.
Our staging method first distinguishes cleavage cycles based on the number or density of nuclei observed. Dorsal membrane morphology is then used to check the assignment of embryos to cleavage cycles C10–14 based on the size and spacing of the nuclei (see Figure 6). Embryos assigned to cleavage cycle C14A are further classified into time classes T1–8 based on membrane morphology (see Figure 7). Using this method, we provide a detailed time-series for expression of the pair-rule gene even-skipped (eve) during the blastoderm stage. Lateral views are shown: enzymatic in situ hybridisation stains to the left, and DAPI-counterstain in the middle. The right-hand column shows details of dorsal membrane/nuclear morphology (sagittal views). See text for details.