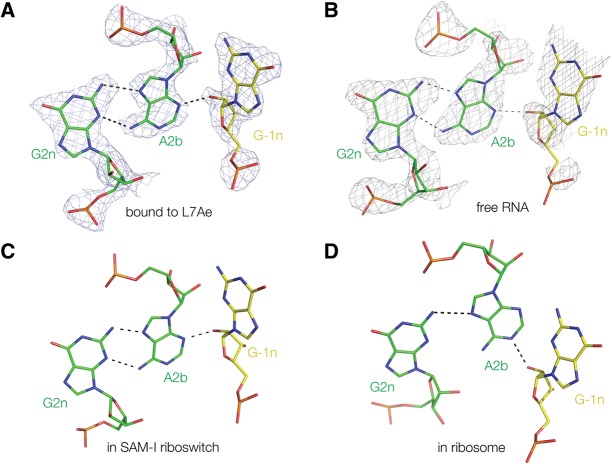
FIGURE 5.
Comparison of the G2n•A2b–A-1n interaction in four Kt-7 structures. (A) Bound by L7Ae protein (this work, with electron density from the composite omit map); (B) as free RNA (this work, with electron density from the composite omit map); (C) engineered into the SAM-I riboswitch (Daldrop and Lilley 2013); and (D) in the H. marismortui ribosomal 50S subunit 23S rRNA (Ban et al. 2000). Structures A–C fall into the N3 class, with G-1n O2′ donating a proton to A2b N3 and two hydrogen bonds connecting A2b and G2n, whereas the ribosomal structure (D) is N1 class where G-1n O2′ donates a proton to A2b N1. The resulting rotation of the A2b nucleobase results in an A2b N6 to G2n N3 N–N length of 4.3 Å, i.e., too long to be hydrogen bonded.