FIGURE 4.
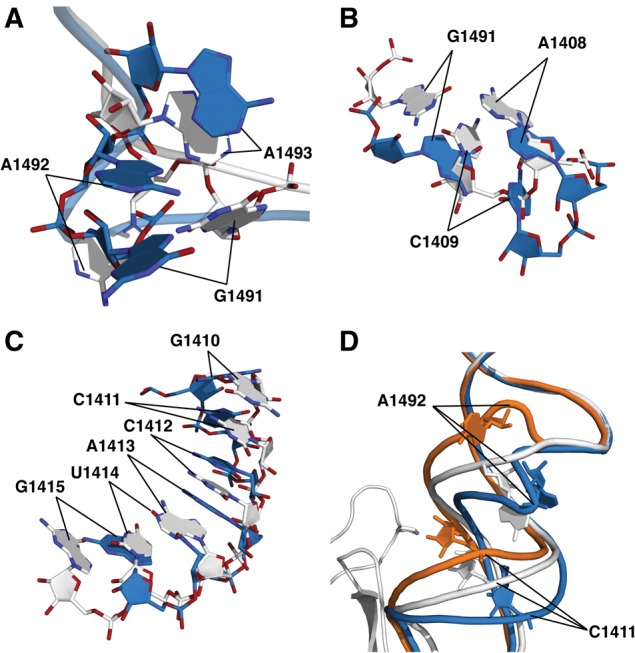
The conformational distortion caused by the SmD mutations. (A) Alignment of wild-type and the P90W mutant in crystal form 2, showing the shifts in position of A1491–A1493. (B) Alignment of wild-type and the P90W mutant in crystal form 2 indicating the shifts of A1408 and the C1409–G1491 base pair. (C) Alignment of wild-type and the P90W mutant in crystal form 2, showing a shift in helix register of bases in the vicinity of G1410–G1415. (D) Alignment of wild-type and P90W mutant in crystal form 2 with the published cocrystal structure of the 30S subunit bound to initiation factor IF1 (PDB entry 1HR0; orange) (Carter et al. 2001). The P90W mutation and IF1 induce shifts centered around A1492 and C1411 that occur in opposite directions. (Gray) The wild-type 30S; (blue) the P90W mutant; (orange) 30S–IF1 complex.
