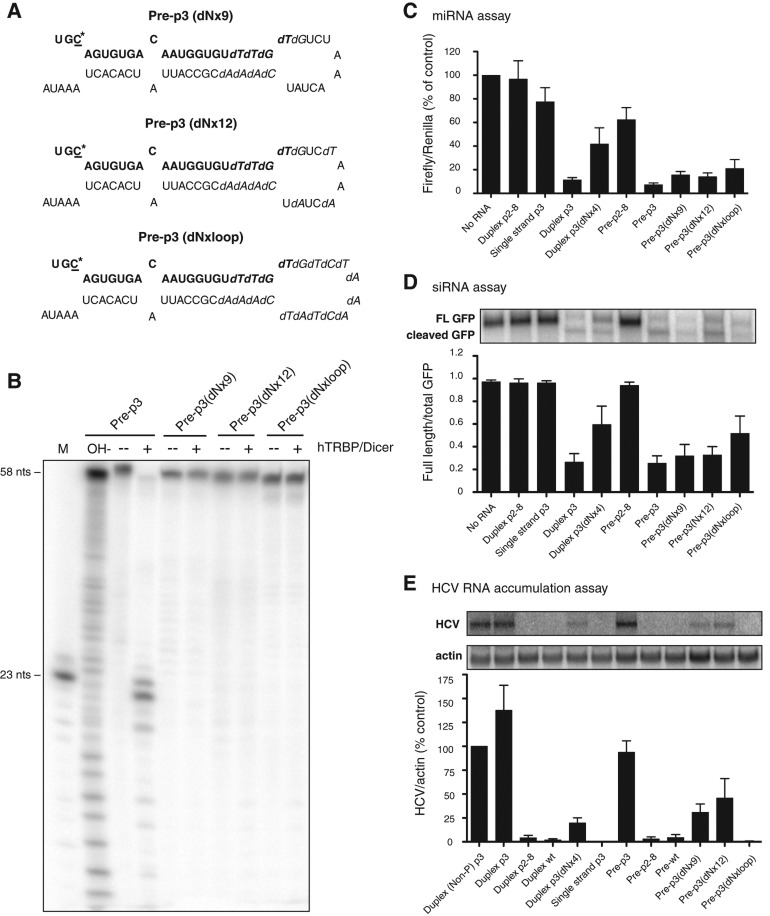
FIGURE 2.
Stabilities and activities of precursor miR-122s containing deoxynucleotides. (A) Sequences and predicted structures of pre-p3 with nine (dNx9), 12 (dNx12), and 19 (dNxloop) deoxynucleotides. The sequence of mature miR-122 is highlighted in bold. The mutated C-nucleotide at position 3 in miR-122 is underlined and marked with an asterisk. Substituted deoxynucleotides are highlighted in italics. (B) hDicer/TRBP cleavage assays of deoxynucleotide-containing precursors. Precursors were labeled at their 5′ ends with 32P and incubated in the presence or absence of purified hDicer/TRBP. Products were separated by gel electrophoresis and exposed to X-ray film. A representative autoradiograph is shown. (C) Luciferase-based microRNA assays. Experiments were performed as in Figure 1C. Data are representative of at least three independent replicates; error bars, SEM. (D) GFP-based siRNA cleavage assays. Various miR-122 species were transfected into cells concurrently with a GFP-expressing plasmid that contains a perfectly complementary site to the mutant miR-122 molecules in its 3′ noncoding region. GFP mRNA was measured by Northern blot analysis. The ratio of full-length GFP mRNA to total (full-length plus cleaved) GFP mRNA was determined, and data from at least three independent replicates are shown. Error bars, SEM. (E) HCV RNA accumulation assays. Rescue of mutated HCV RNA by miR-122 species, carrying compensatory mutations, after transfection into Huh-7 cells. RNA accumulation was measured by Northern blot analysis. Autoradiographs were quantitated using ImageQuant, and HCV RNA amounts were normalized to that of γ-actin RNA. Data from cells transfected with p3 were set to 100% and used as a positive control. The data shown represent at least three independent replicates. Error bars, SEM.