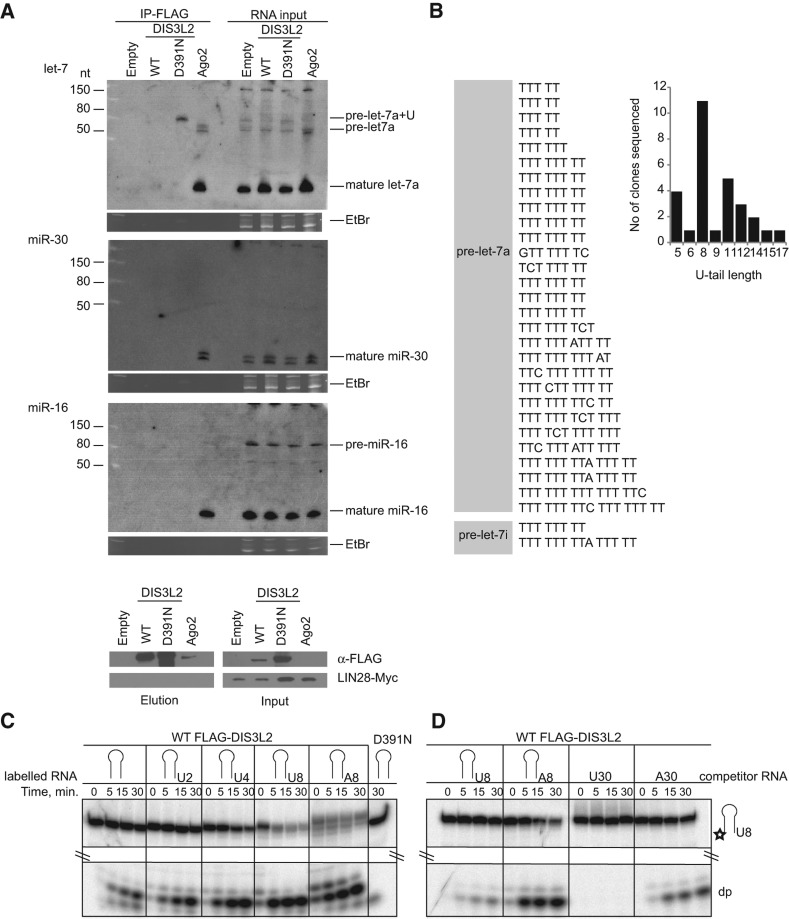
FIGURE 3.
DIS3L2 targets 3′ end-uridylated precursors of let-7 miRNA. (A) Mutant DIS3L2 (D391N) specifically coprecipitates with extended forms of pre-let-7a miRNA. Northern blot analyses of RNAs coimmunoprecipitated with FLAG-tagged proteins as indicated on the upper side of the autoradiograph (IP-FLAG). The RNA input was total RNA isolated from whole cell lysates. The ethidium bromide (EtBr)-stained gels represent input loading controls. (Empty) Control RIP performed using cells that were not expressing any FLAG-tagged protein. The lower panel shows Western blot analysis of Myc-tagged LIN28A expression and the efficiency of FLAG tag-mediated immunoprecipitation of proteins used for RIP analyses. “Input” shows the whole cell lysates used for the IPs. The ectopic expression of Myc-tagged LIN28A was monitored with specific anti-Myc antibodies. DIS3L2 was detected with anti-FLAG antibodies. (B) Sequencing analysis of the 3′ termini of pre-let-7 RNAs coprecipitated with the D391N DIS3L2 mutant in HEK293T-Rex cells ectopically overexpressing LIN28A and pri-let-7a-1. The analysis suggests that the DIS3L2 mutant binds to uridylated pre-let-7a-1 and pre-let-7i because 31 out of 35 sequenced clones contained nontemplate oligo(U)-tails. The gray box schematically represents the body of pre-let-7 miRNAs. A graphical representation of the (U)-extension statistics is shown at the right. (C) Oligouridylation stimulates DIS3L2-mediated degradation of pre-let-7 miRNA. An in vitro degradation assay with FLAG-DIS3L2 purified from HEK293T-Rex cells and pre-let-7 RNA (represented by the schematic stem–loop) containing either no 3′ extension or 3′ oligo(U) and oligo(A) extensions of increasing lengths (indicated at the top) as substrates. D391N indicates a reaction with purified catalytically inactive DIS3L2. (D) DIS3L2 exhibits a preference for RNA substrates with oligo(U) extensions. A 5′ end-labeled pre-let-7-U8 RNA was incubated with purified DIS3L2 in the presence of a 10-fold excess of different unlabeled competitor RNAs (indicated at the top) for the time periods shown.