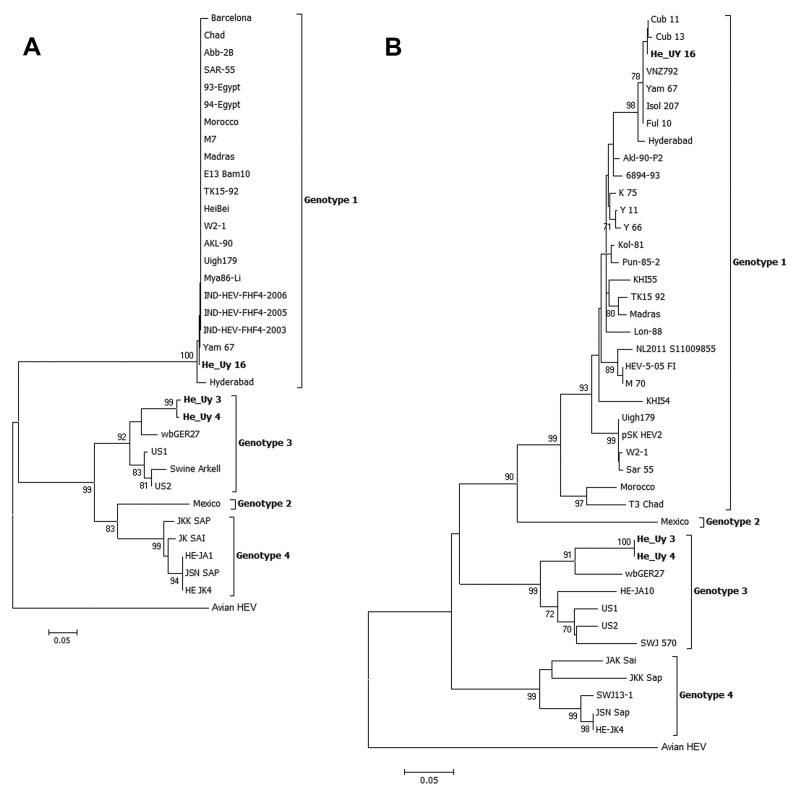
Figure.
Phylogenetic tree of the Hepatitis E virus constructed on the basis of A) the partial 137-nt open reading frame (ORF) 2–3 overlapping region and B) the 244-nt sequence of the RNA dependent RNA polymerase within ORF1. Trees were generated by using the neighbor-joining method with the Kimura 2-parameter as the substitution model. The robustness of the trees was determined by bootstrap for 1,000 replicates. Values ≥70% are shown. Strains isolated in Uruguay are shown in boldface. The isolate of genotype 1 detected in this study (He_Uy 16) clustered (98% bootstrap value) with genotype 1 strains from India and Latin America (Cub 11 and 13 and VNZ792). Scale bars indicate nucleotide substitutions per site.