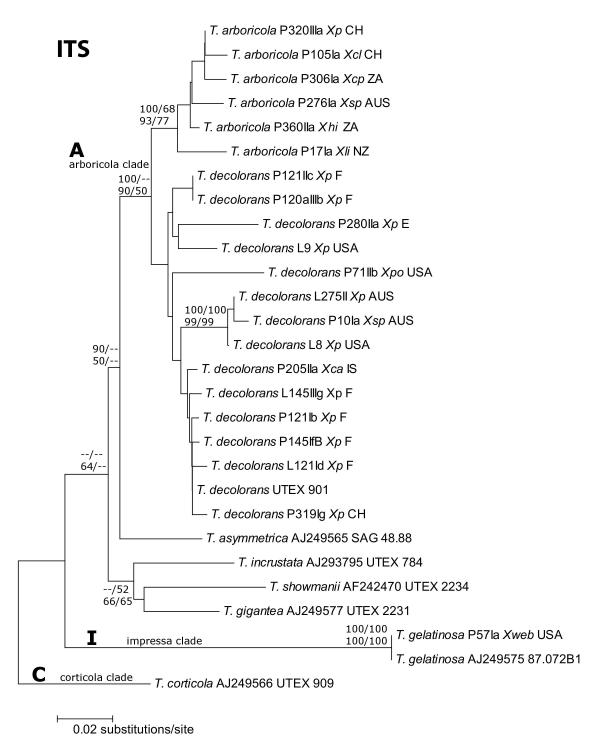
Fig. 1.
Phylogeny of Trebouxia s. lat.. Sequences obtained from GenBank are indicated in Table 3. Support values shown at a respective node are posterior probabilities of nodes from Bayesian analysis in MrBayes (first row, first number), values from 100 bootstrap replicates in a maximum likelihood analysis in Paup (first row, second number), values from 500 bootstrap replicates in a neighbor-joining analysis in MEGA (second row, first number) and maximum parsimony support values from 100 bootstraps generated with PAUP (second row, second number). Clades A, I and C represent ‘arboricola’, ‘impressa’ and ‘corticola’, respectively, as proposed by Helms (2003). The identity of photobionts was inferred on the basis of ITS and rbcL phylogenetic analyses including reference strains (Nyati, Scherrer, Werth and Honegger, in press). Abbreviations used: Xca: Xanthoria candelaria, Xcl: X. calcicola, Xcp: X. capensis, Xec: X. ectaneoides, Xh: X. hirsuta, Xli: X. ligulata, Xp: X. parietina, Xpo: X. polycarpa, Xtu: X. turbinata, Xsp: Xanthoria sp., Xweb: Xanthomendoza weberi.. Phylogeny of the nuclear ribosomal internal transcribed spacer region (complete ITS region, including the 5.8S rRNA gene) as inferred using Maximum Likelihood (ML) analysis. Asterochloris sequences not used in analyses due to very high sequence divergence.