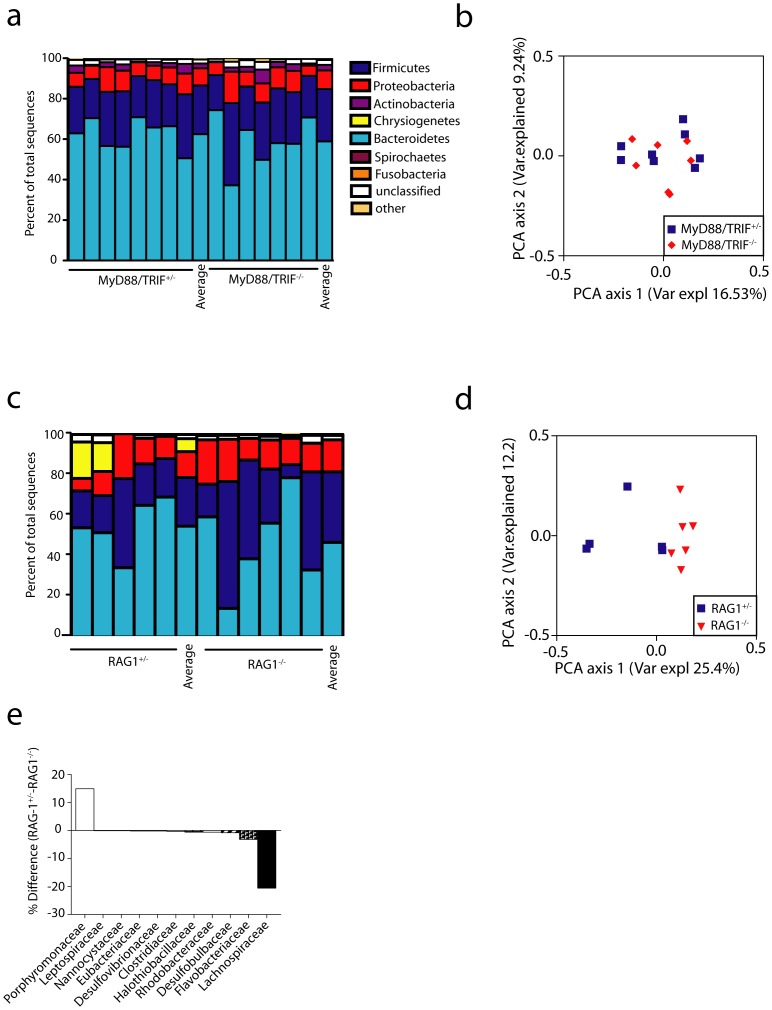
Figure 4. The colonic microbiome is altered in Rag1−/− but not MyD88/TRIF−/− mice.
(a) Phyla distribution and (b) PCoA analysis (based on unweighted UniFrac distances among samples) of microbiomes from fecal pellets of MyD88/TRIF+/− (n = 8) and MyD88/TRIF−/− (n = 7) mice is shown. No significant differences based on ANOSIM were observed (c) Phyla distribution and (d) PCoA analysis from Rag1−/− (n = 6) and Rag1+/− (n = 5) fecal pellets is shown. Community composition differs between Rag1+/− and Rag1−/− based on ANOSIM, p = 0.004. (e) Comparison of bacterial family abundances between RAG1−/− and RAG1+/−, differences are shown in percent.