Figure 4. Meiotic double-strand breaks are responsible for the elevated cell death observed following errors in mitosis.
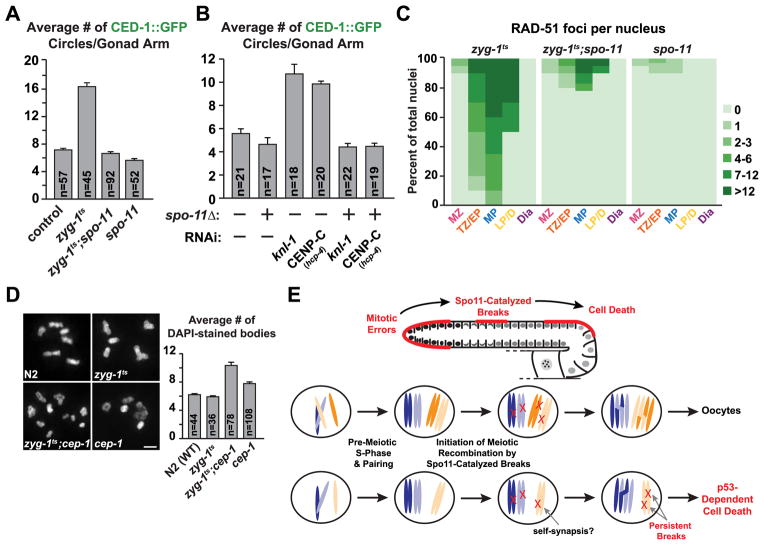
(A) Quantification of cell death following inhibition of spo-11 in both control and zyg-1ts 24h post upshift. Error bars are SEM.
(B) Quantification of cell death following inhibition of spo-11 in both control and indicated mitotic inhibitions (experimental scheme, Fig. 1B). Error bars are SEM.
(C) Quantification of the RAD-51 foci for the indicated genotypes 48h post upshift (zyg-1ts values are the same as in Fig. 3F).
(D) Representative images and quantification of the chromosome content of oocytes for control, zyg-1ts, zyg-1ts;cep-1 and cep-1 72h post upshift. Errors bars are SEM; unpaired t test shows high significance (p<0.0001) between zyg-1ts and the double mutant. Bar, 2 μm.
(E) Schematic summarizing the role of Spo11-catalyzed breaks in the detection of mitotic errors that occurred in the distal gonad. The individual nucleus cartoons below show a normal nucleus with 2 pairs of homologous chromosomes (top) and a defective nucleus in which one of the chromosomes is missing due to missegregation (bottom). For simplicity, potential self-synapsis that would result in accumulation of DNA double-strand breaks is suggested but not depicted in the cartoon.
(See also Figure S4).