Figure 6. The clade-specific sites in the MAGO and Y14 families.
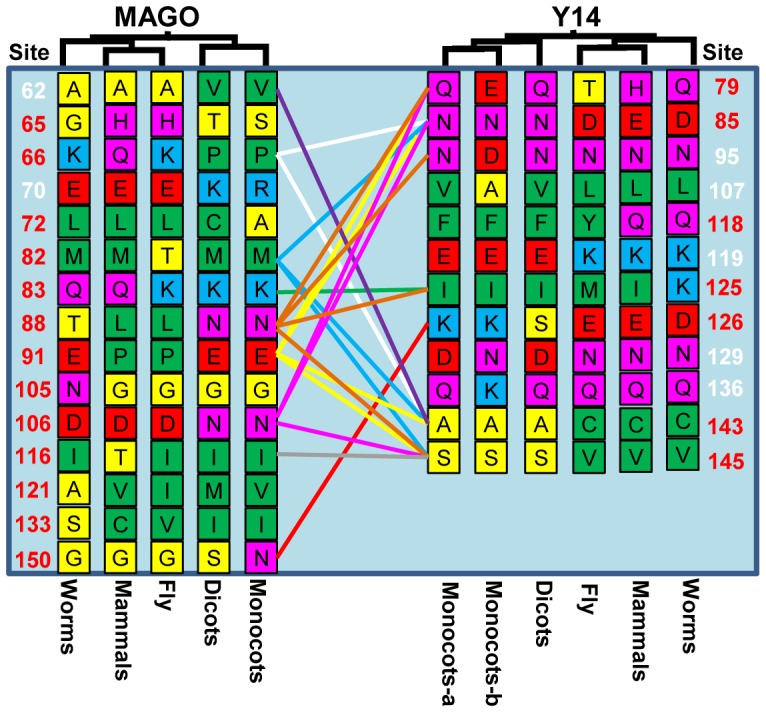
The clade-specific amino acids are arranged that the small nonpolar residues (G, A, S and T) are highlighted in yellow, the hydrophobic residues (C, V, I, L, P, F, Y, M and W) in green, the polar residues (N, Q and H) in magenta, the negatively charged residues (D and E) in red, and the positively charged residues (K and R) in blue. The residues between the two protein families that were predicted to be correlated mutation groups in CAPS were connected by color lines. The black, red, green, blue, yellow, purple, pink, orange and gray line respectively represents the G4, G5, G6, G12, G18, G24, G33, G35 and G40 (Figure 5; Table S5 in File S1). The given position of the residues in the MAGO and Y14 families corresponds to OsMAGO2 and OsY14b, respectively. The red numbers showed that these sites were predicted to be co-evolved in CAPS, the white was not.
