Abstract
Autosomal Recessive Osteopetrosis is a genetic disorder characterized by increased bone density due to lack of resorption by the osteoclasts. Genetic studies have widely unraveled the molecular basis of the most severe forms, while cases of intermediate severity are more difficult to characterize, probably because of a large heterogeneity. Here, we describe the use of exome sequencing in the molecular diagnosis of 2 siblings initially thought to be affected by “intermediate osteopetrosis”, which identified a homozygous mutation in the CTSK gene. Prompted by this finding, we tested by Sanger sequencing 25 additional patients addressed to us for recessive osteopetrosis and found CTSK mutations in 4 of them. In retrospect, their clinical and radiographic features were found to be compatible with, but not typical for, Pycnodysostosis. We sought to identify modifier genes that might have played a role in the clinical manifestation of the disease in these patients, but our results were not informative. In conclusion, we underline the difficulties of differential diagnosis in some patients whose clinical appearance does not fit the classical malignant or benign picture and recommend that CTSK gene be included in the molecular diagnosis of high bone density conditions.
Keywords: Exome sequencing, CTSK, Sclerosing bone disorder, Differential diagnosis, Therapy
Highlights
-
•
Whole exome sequencing was applied to 2 patients with “intermediate osteopetrosis” and negative mutation screening in ARO genes.
-
•
We identified CTSK gene mutations in these as well as in additional patients referred with “intermediate osteopetrosis”.
-
•
We investigate the role of modifier genes possibly impacting on the clinical manifestation of the disease in these patients.
-
•
We recommend that CTSK gene analysis be included in the molecular workup of high bone density conditions.
Introduction
The osteopetroses are a group of clinically and genetically heterogeneous bone diseases sharing the hallmark of increased bone density on radiographs [1]. This pathological feature results from abnormalities in either osteoclast differentiation or function [2]. Clinical and molecular dissection of osteopetroses has identified forms with different severity and prognosis [3], even though classification of single patients into a specific subgroup is not always easy due to the rareness of these conditions and to the presence of a variety of additional clinical features. On the other hand, the possibility to obtain a precise molecular diagnosis importantly impacts on the patients' management [2], [3].
Since its first application few years ago [4], [5], [6], whole exome sequencing has been exploited to identify the causative gene of many monogenic disorders, including skeletal diseases. Exome sequencing offers the great advantage that large groups of affected individuals are not required; on the contrary, the analysis can be successfully conducted in a single family, which is very useful when only atypical or extremely rare forms of a heterogeneous disease are left for diagnosis.
Here we report the results of exome sequencing in 2 siblings with an initial clinical diagnosis of intermediate osteopetrosis, which identified a mutation in the Cathepsin K (CTSK) gene, known to cause Pycnodysostosis (MIM 265800). This finding prompted us to analyze the same gene in 25 patients addressed to us with a clinical diagnosis of recessive osteopetrosis with no recognized genotype, leading to the identification of mutations in CTSK in 4 additional patients.
The cathepsins constitute a family of lysosomal cysteine proteases responsible for several important cellular processes [7]. They are produced in an inactive form containing an N-terminal proregion, which is cleaved upon activation and required for proper protein folding and intracellular trafficking, and for inhibition of the proteolytic function until the proenzyme reaches the lysosome [8]. In particular, Cathepsin K is a marker of late osteoclast differentiation with an important role in the degradation of bone organic matrix [9]. In addition, studies in animal models demonstrated its involvement in autoimmune and inflammatory diseases through regulation of Toll-like receptor 9 (TLR9) signaling [10].
Our molecular results allowed redirecting the clinical diagnosis in 6 patients, in support of the possibility that exome sequencing is routinely used as a diagnostic tool in the near future, especially for disorders that share a common clinical presentation but are genetically heterogeneous.
Materials and methods
Samples
Clinical data and specimens, including blood and DNA samples, were collected from patients and their parents after informed consent. This research complies with the standards established by the Independent Ethical Committee of the Humanitas Clinical and Research Centre.
Exome analysis
Exome capture was performed using 1.5 μg of high-quality genomic DNA from each patient and the TruSeq Exome Enrichment Kit (Illumina) that provides coverage across 62 Mb of exomic sequence, including 5′ UTR, 3′ UTR, microRNA and other non-coding regions. The enriched library was validated by the Agilent DNA 1000 Kit and loaded on the cBot Station (Illumina) to create clonal clusters on the flow cell. Sequencing was performed at the CRS4 center (Centro di Ricerca, Sviluppo e Studi Superiori in Sardegna, Italy) on the Hiseq2000 Instrument. Reads extracted with the Illumina tools were aligned to the reference genome hg19 by using Seal 0.2.3 and stored in compressed binary files (BAM). Single nucleotide variations as well as insertions and deletions were called using the Genome Analysis Toolkit (GATK) [11]. Quality controls were performed using the QC Tool [12]. The variants thus identified were merged in a unique list, in a Variant Call Format (VCF) file, and successively annotated by ANNOtation of genetic VARiants (ANNOVAR) [13]. Finally, variants were further prioritized and filtered according to a basic workflow for exome sequencing.
Sanger sequencing
CTSK gene amplification and direct sequencing of exons and intron–exon boundaries were performed as described [14]. The mutation nomenclature conforms to HGVS (www.hgvs.org/mutnomen) [15]; the reference sequence for the genomic DNA is GenBank NC_000001.10, while for the cDNA is GenBank NM_000396.2 (the numbering starts with nucleotide + 1 for the A of the ATG-translation initiation codon).
Primer sequences and conditions for amplification and sequencing of selected genomic regions of Low density lipoprotein receptor-related protein 4 (LRP4), Filamin B (FLNB), Cerberus 1 homolog (CER1) and Osteopontin (OPTN) genes are available upon request.
In silico analysis
The putative effect of the mutations identified in CTSK gene was predicted using the publicly available tools Mutation Taster (http://www.mutationtaster.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT and Provean (http://provean.jcvi.org/genome_submit.php).
Results
Patients
Family 1 came from Kashmir (Pakistan) and comprised 2 affected siblings born from consanguineous parents (first cousins). Both patients were reported to have a “slow onset” form of osteopetrosis which was thought to be of autosomal recessive inheritance due to parental consanguinity and the absence of symptoms possibly related to the disease in their parents. The elder child (Patient 1A) had a transient anemia as an infant, but since has had normal blood counts (Hb 12 g/dl, WBC 10.1 × 109/l, neutrophils 2.37 × 109/l and platelets 255 × 109/l, at 12 years). Growth retardation was reported and became more striking with the age (height < 0.4th centile and weight at 0.4th centile) at 12 years. She presented with proptosis and became totally blind when she was 5 years old. At 11 years she had an episode of dysesthesia in both arms and legs and an MRI examination showed a Chiari malformation. An intra-ventricular shunt was inserted to reduce intracranial pressure. At the moment she is not receiving any therapy, attending school in reasonably good conditions.
Her younger sister (Patient 1B) was diagnosed in the first year of life due to family history, and showed mild anemia and short stature (height < 3rd centile at 34 months). Diagnosis was confirmed by plain X-rays. When she was 2.5 years old, she began to display visual impairment, despite normal visual evoked potentials (VEP). Bronchiectasis was also present. At 3 years she received matched bone marrow transplantation (BMT) after conditioning according to the European Group for Bone Marrow Transplantation-European Society for Immunodeficiencies (EBMT-ESID) guidelines (www.esid.org/downloads/OPGuidelines-2011). She reached full engraftment, even though bone improvement was evident only after 5 months; post-transplant complications were graft versus host disease (GvHD), grade 1, and transient mild veno-occlusive disease (VOD). Ophthalmological follow-up showed slightly reduced visual acuity (right − 0.14 and left 0.14), full visual fields and pink optic nerves. Growth is poor at 8.5 years even though partially improved post-BMT (height from 0.3 cm below the 0.4th centile before BMT to 0.1 cm below the 0.4th centile). Hematological parameters are now within normal range.
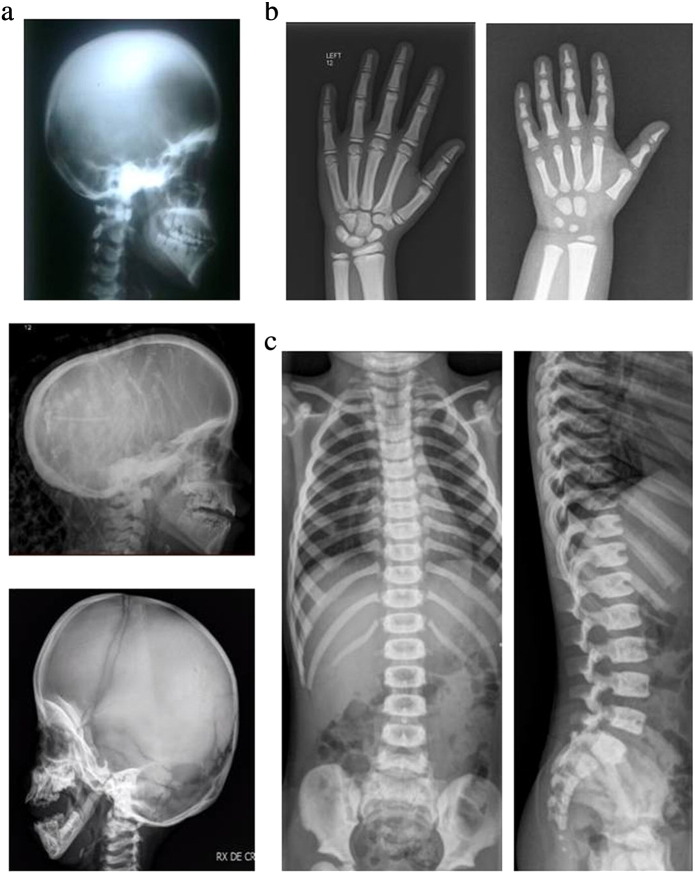
Patient 2 was born from consanguineous Pakistani parents (first cousins). She presented with the complaint of progressive pallor for one month at 12 years of age. On examination she showed severe anemia (Hb 6.3 g/dl) and splenomegaly, raising the suspicion of hemolytic anemia; however, work up turned out to be negative. The presence of growth retardation (height < 2nd centile, weight 9th centile) and complete skeletal survey led to the diagnosis of osteopetrosis (see Fig. 1a upper panel, for the most recent radiological cranial evaluation). She was initially treated with steroids and calcitriol and then received blood transfusion from the age of 15; at present (17 years old) she is on calcitriol only. She presents proptosis, malar prominence and short stature.
Fig. 1.
Selected radiographs of Patient 2 at 17 years of age, Patient 4 at 8 years of age and Patient 5 at 4 years of age. A diffuse increased bone density is evident. Panel a: Lateral view of the skull (Patient 2 top; Patient 4 middle; Patient 5 bottom). Loss of the mandible angle is present only in Patient 5, which also presents wormian bones. The superposition on the X-ray in the middle is caused by braids (the patient being of Caribbean origin). Panel b: Hand radiographs of Patient 4 (left) and Patient 5 (right). The distal phalangeal tufts are small and this might be considered an early sign of acroosteolysis (but also a normal variant); however no overt signs of acroosteolysis are present, possibly because of their relatively young age. Only the diaphyseal constriction of metacarpals in Patient 5 is suggestive of Pycnodysostosis. Panel c: “Sandwich vertebrae” in Patient 5. The lateral view shows peripheral bone sclerosis with central hypo-densities and a wide anterior notch corresponding to the arterial canal.
Patient 3 was born from consanguineous Bangladeshi parents. He was diagnosed with mild osteopetrosis at 9 months due to a generalized increase in bone density on X-ray and visual impairment requiring optic nerve decompression (at 9 years of age), while the hematological compartment was normal. He has had also recurrent mal-uniting fractures of the femur. At present he is alive and clinically stable at 19 years of age.
Patient 4 was born from Black Caribbean unrelated parents. She was accidentally diagnosed at 3 years of age, during a routine X-ray performed after swallowing a screw. She also displayed moderate anemia (Hb 10.4 g/dl) and mild visual impairment with a slight nystagmus, while on a CT scan foramen magnum narrowing and a syrinx were present. At the age of 7 she underwent a foramen magnum decompression for cerebellar tonsil ectopia and developed hydrocephalus in the postoperative period requiring placement of ventriculo-peritoneal shunt. She is alive at 10 years of age with stable hematological conditions, an important syrinx in the spinal cord, and obstructive sleep apnea requiring nocturnal continuous positive airway pressure. The available X-rays also show scaphocephaly (Fig. 1a central panel), which is rarely seen in osteopetrosis while it has been reported in Pycnodysostosis; distal phalangeal tufts are small, but no overt signs of acroosteolysis are apparent (Fig. 1b left panel).
Patient 5 was born from Pakistani, reportedly unrelated parents. Since the age of 3, he was followed due to growth retardation (height < 3rd centile at 5 years of age) and anemia (Hb 8.8 g/dl). Recently, skeletal survey showed the presence of osteopetrotic radiological signs including generalized increase in bone density (Fig. 1b right panel), cranial sclerosis particularly at the skull base (Fig. 1a bottom), widening of anterior ribs and long bone metaphysis, Erlenmeyer flask deformity of distal femurs and suggested "sandwich vertebrae" (Fig. 1c). However, the loss of the mandible angle and the presence of wormian bones might have suggested a diagnosis of Pycnodysostosis (Fig. 1a bottom). He is alive at 5 years in reasonably good conditions.
In all patients laboratory findings regarding the immune compartment were within a normal range, even though no extensive characterization was done.
Molecular results
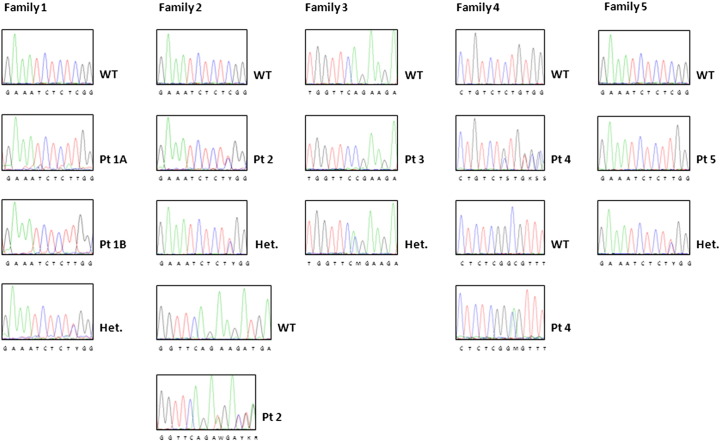
We performed exome sequencing in the 2 affected siblings of Family 1 and achieved in both patients a 69 × mean coverage over the 62 Mb targeted exome, with more than 94% of targeted regions covered. The overall transition to transversion rate (Ti/Tv) was 2.50 in line with what was expected for exome sequencing. The analysis identified a total of 179143 variants which were filtered with dbSNP137 and 1000 Genome Project and according to the pattern of inheritance of the disease and to the parental consanguinity (Table 1). Among the homozygous variants, we found a mutation in exon 3 of the CTSK gene (g.2128C > T) which could be considered responsible for the disease in Patients 1A and 1B (Table 2); of note, the same mutation, leading to an amino acid substitution at codon 46 (p.Arg46Trp), was already known to cause Pycnodysostosis [16]. The nucleotide change was confirmed by Sanger sequencing in the homozygous state in the patients and in the heterozygous state in their parents (Supplementary Fig. 1, which also shows the mutations found in the other patients).
Table 1.
Number of variants/genes identified in patients 1A and 1B through exome sequencing.
| Total variants | 179143 |
| Shared variants after base quality filtering | 126965 |
| Homozygous variants | 16082 |
| Non-synonymous/indel/splice site variants | 3525 |
| Novel variants (dbSNP137/1000GP queried) | 165 |
| Genes with plausible disease association | 3 |
Table 2.
Molecular findings in 6 new CTSK-dependent patients.
| Patient | Genomic changea | Location in DNA | cDNA changeb | Protein change/effect | Location in protein |
|---|---|---|---|---|---|
| 1A and 1B | g.2128C > T g.2128C > T |
Exon 3 Exon 3 |
c.136C > T c.136C > T |
p.Arg46Trp p.Arg46Trp |
Proregion |
| 2 | g.2128C > T g.2343_2345del |
Exon 3 Exon 4 |
c.136C > T c.266_268del |
p.Arg46Trp p.Lys89del |
Proregion |
| 3 | g.2340A > C g.2340A > C |
Exon 4 Exon 4 |
c.263A > C c.263A > C |
p.Gln88Pro p.Gln88Pro |
Proregion |
| 4 | g.2131C > A g.8746_8747del |
Exon 3 Exon 6 |
c.139C > A c.737_8delCT |
p.Arg47 Ser p.Ser246CysfsX4 |
Proregion Mature protein |
| 5 | g.2128C > T g.2128C > T |
Exon 3 Exon 3 |
c.136C > T c.136C > T |
p.Arg46Trp p.Arg46Trp |
Proregion |
Accession number genomic sequence of the CTSK gene: NC_000001.10.
Accession number of the CTSK cDNA: NM_000396.2; the numbering used starts with nucleotide + 1 for the A of the ATG-translation initiation codon.
Supplementary Fig. 1.
Molecular findings in 6 new CTSK-dependent patients.
Family 1: A homozygous single nucleotide change (g.2128C > T) causing an Arg46Trp change was identified in the two affected siblings; this mutation was present in the parents at the heterozygous state (see lowest chromatogram in this series).
Family 2: Patient 2 was a compound heterozygote for the nucleotide change above described (present also in the paternal DNA at the heterozygous state) and a deletion of 3 nucleotides (g.2343_2345del), leading to the deletion of a single residue (p.Lys89del).
Family 3: A homozygous transversion (g.2340A > C) causing a Gln88Pro change was identified in the patient; this mutation was present in the parents at the heterozygous state (see lowest chromatogram).
Family 4: Patient 4 was a compound heterozygote for a single nucleotide (g.2131C > A), causing an Arg47Ser substitution, and a deletion of 2 nucleotides (g.8746_8747del), causing a frameshift and a premature protein termination (p.Ser246CysfsX4).
Family 5: The same single nucleotide change found in the patients 1A, 1B and 2 was present at the homozygous state in Patient 5 and at the heterozygous state in his parents (see lowest chromatogram in this series).
This finding prompted us to sequence the CTSK gene in other 25 patients sent us with a clinical diagnosis of autosomal recessive osteopetrosis (ARO) but in whom we could not identify a molecular defect in the known ARO genes [3]. Among these patients we identified 4 individuals bearing mutations in the CTSK gene. In particular, Patient 2 was a compound heterozygote for the nucleotide change above described and a deletion of 3 nucleotides in exon 4 (g.2343_2345del), leading to the deletion of a single residue (p.Lys89del). Her father was heterozygous for the missense mutation, while maternal DNA was not available as the patient's mother deceased several years earlier.
Patient 3 was homozygous for a transversion in exon 4 (g.2340A > C) leading to an amino acid substitution at codon 88 (p.Gln88Pro); this nucleotide change was confirmed in her parents in the heterozygous state.
Patient 4 was compound heterozygous for a nucleotide change in exon 3 (g.2131C > A), causing an amino acid substitution at codon 47 (p.Arg47Ser), and a deletion of 2 nucleotides in exon 6 (g.8746_8747del), causing a frameshift and a premature protein termination (p.Ser246CysfsX4).
Patient 5 was homozygous for the same nucleotide change found in patients 1A, 1B and 2 (g.2128C > T); his parents carried this mutation in the heterozygous state.
Apart from p.Arg46Trp, the other changes are herein described for the first time. The 3 missense mutations (p.Arg46Trp, p.Arg47Ser and p.Gln88Pro) and the single amino acid deletion (p.Lys89del) are located in the CTSK proregion, while the frameshift (p.Ser246CysfsX4) affects the mature enzyme.
As already reported for Arg46, the neighboring Arg47 is highly conserved among species and is also found in the corresponding position in human cathepsins K, S and L [16].
The missense substitutions (p.Arg46Trp, p.Arg47Ser and p.Gln88Pro) and the single amino acid deletion (p.Lys89del) were not found in more than 100 chromosomes from healthy unrelated individuals from the same geographical area, and were not present in SNP databases; therefore they are unlikely to be neutral polymorphisms. Of note, in silico analysis using several tools (Mutation Taster, PolyPhen-2, SIFT, Provean) predicted a damaging effect for all of them.
In addition, exome sequencing data in the affected siblings of Family 1 detected a number of known both homozygous and heterozygous single nucleotide variants (SNV) in a set of genes already associated with bone defects or bone mineral density (Supplementary Table 1). In this list, we selected exonic non-synonymous SNVs with a minor allele frequency below 0.1 in both the Exome Sequencing Project (ESP6500) and the 1000 Genome Project; this value was chosen based on the hypothesis that variants less frequent in the general population might more importantly impact on the disease-causing allele. We speculated that the presence of one or more specific SNVs in all the patients here described could modify the classical pycnodysostotic phenotype. So, we genotyped the selected variants in all six patients, but we could not identify a shared genotype or SNV (Supplementary Table 2).
Discussion
To date, the molecular and cellular basis of a considerable number of genetic disorders is still unknown and this knowledge gap is reflected in not always satisfactory diagnostic and therapeutic strategies. However, the contribution of new, high-throughput techniques for the sequencing of the human genome has importantly speeded up the identification of the genes responsible for many diseases. In particular exome sequencing has come to the fore only few years ago, but has already widely demonstrated its power in identifying both new disease genes and new genotype–phenotype associations [17]. Our results further support the role of exome sequencing in the differential diagnosis of genetically heterogeneous diseases. The clinical presentation of 6 patients in our cohort was originally described as mild osteopetrosis, but molecular analysis failed to detect mutations in any of the genes known to cause this phenotype in humans. Exome sequencing in 2 affected siblings detected a mutation in the CTSK gene already reported in Pycnodysostosis [16], and mutations in the same gene were subsequently found in the remaining 4 affected individuals. Pycnodysostosis shares with ARO some clinical features, such as a generalized increase in bone density, frontal bossing, short stature, delayed abnormal tooth eruption and fragility fractures. Indeed, one of the 2 original descriptions of Pycnodysostosis in 1962 named it an osteopetrosis variant [18], [19]. However, Pycnodysostosis is usually a progressive but relatively benign condition. It presents in the first years of life with short stature, a peculiar facial appearance with bi-temporal narrowing, and clinical and radiographic signs such as stubby hands and feet with acroosteolysis, hypoplasia of the maxilla and absence of the mandible angle, which are considered essentially pathognomonic [20], [21]. Evaluation of the radiographic documentation available from 3 out of 6 patients showed in 2 of them absence of the obtuse mandible angle on a craniolateral view, and in all of them absence of obvious acroosteolysis of the hands, thus suggesting that the radiological evidence was not sufficient for an unequivocal clinical classification.
Variants in other genes involved in bone homeostasis might have impacted on the radiological presentation of these patients. Proving which variants are actually playing as modifiers of a given condition is not a trivial issue [22]. In the present work, we restricted the analysis to coding, non-synonymous SNV with low frequency in the general population, found in genes related to bone phenotypes; however, this strategy did not identify a genotype common to all the patients, which could support the idea of an involvement in disease modulation. A more comprehensive study including also synonymous and non-coding variants, genotyping of a larger cohort of patients and functional studies might have more chances to succeed, but in our case it could not be performed due to the limited sample size.
Overall, our results show that, when the defects commonly referred to as pathognomonic of a specific skeletal disease are absent or are not evaluated correctly, the radiographic signs of increased bone density can be non-specific and insufficient to point at a specific diagnosis, as occurred in our patients. In this case, the genetic analysis becomes crucial. Indeed, several investigators who have applied whole exome sequencing in the clinical diagnostics have remarked that so-called “atypical” or incomplete cases that do not fulfill the textbook diagnostic criteria seem to be common [23], [24]. In other words, atypical patients must be much more frequent than hitherto appreciated. This is a strong point in favor of a broader and unbiased approach to molecular diagnostics.
Exploiting new sequencing technologies, a “gene panel” approach can be implemented in the diagnosis of conditions that share clinical signs but have a heterogeneous molecular basis (e.g., lysosomal storage diseases with skeletal involvement or osteogenesis imperfecta and bone fragility disorders, known to be associated with more than 10 different genes) [1]. Indeed different platforms designed to enrich the target regions of genes implicated in specific bone diseases are under development as rapid and powerful diagnostic tools [25]. An alternative strategy could consist in limiting the analysis of data from whole exome sequencing to only the genes relevant to the patient phenotype; this would allow to increase the number of genes under investigation without repeating the test, in case the first selected subpanel is negative [26].
In conclusion, although a careful examination of the clinical picture and of high quality X-rays might correctly have raised the right diagnostic suspicion in some of our cases, thus avoiding the decision to use the exome approach, we would recommend that CTSK gene be included in the molecular diagnosis of intermediate forms of human ARO and, more in general, of high-density bone conditions, even when Sanger sequencing is used for the mutation screening.
The following are the supplementary data related to this article.
List of genes associated to bone phenotypes.
Genotype of selected, putative modifier genes in our 6 CTSK-dependent patients.
Disclosure statement
The authors have nothing to disclose.
Acknowledgments
This work was partially supported by the Telethon Foundation [grant GGP12178 to C.S.]; by PRIN Project [200999KRFW-002 to P.V. and 20102M7T8X_003 to A.V.]; by Giovani Ricercatori from Ministero della Salute [grant GR-2008-1134625 to C.S.]; by Ricerca Finalizzata from Ministero della salute [RF-2009-1499,542 to A. Villa] and by PNR-CNR aging Program 2012–2014.
Edited by: Bente Langdahl
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Warman M.L., Cormier-Daire V., Hall C., Krakow D., Lachman R., LeMerrer M. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943–968. doi: 10.1002/ajmg.a.33909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villa A., Guerrini M.M., Cassani B., Pangrazio A., Sobacchi C. Infantile malignant, autosomal recessive osteopetrosis: the rich and the poor. Calcif Tissue Int. 2009;84:1–12. doi: 10.1007/s00223-008-9196-4. [DOI] [PubMed] [Google Scholar]
- 3.Sobacchi C., Schulz A., Coxon F.P., Villa A., Helfrich M.H. Human osteopetrosis: genetics, treatment and new insights into osteoclast function. Nat Rev Endocrinol. 2013;9:522–536. doi: 10.1038/nrendo.2013.137. [DOI] [PubMed] [Google Scholar]
- 4.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoischen A., van Bon B.W., Gilissen C., Arts P., van Lier B., Steehouwer M. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 6.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M. Exome sequencing identifies the cause of a Mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turk V., Turk B., Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cygler M., Mort J.S. Proregion structure of members of the papain superfamily. Mode of inhibition of enzymatic activity. Biochimie. 1997;79:645–652. doi: 10.1016/s0300-9084(97)83497-9. [DOI] [PubMed] [Google Scholar]
- 9.Inui T., Ishibashi O., Inaoka T., Origane Y., Kumegawa M., Kokubo T. Cathepsin K antisense oligodeoxynucleotide inhibits osteoclastic bone resorption. J Biol Chem. 1997;272:8109–8112. doi: 10.1074/jbc.272.13.8109. [DOI] [PubMed] [Google Scholar]
- 10.Asagiri M., Hirai T., Kunigami T., Kamano S., Gober H.J., Okamoto K. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- 11.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berutti R., Reinier F., Atzeni R., Angius A., Cusano R., Marcelli M. ESHG; Nürnberg, Germany: June 23–26, 2012. QCTool: an efficient toolkit to automatically generate quality metrics of next-generation sequencing data. [P11.119] [Google Scholar]
- 13.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnarumma M., Regis S., Tappino B., Rosano C., Assereto S., Corsolini F. Molecular analysis and characterization of nine novel CTSK mutations in twelve patients affected by pycnodysostosis. Hum Mutat. 2007;28:524–533. doi: 10.1002/humu.9490. [DOI] [PubMed] [Google Scholar]
- 15.den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 16.Schilling A.F., Mülhausen C., Lehmann W., Santer R., Schinke T., Rueger J.M. High bone mineral density in pycnodysostotic patients with a novel mutation in the propeptide of cathepsin K. Osteoporos Int. 2007;18:659–669. doi: 10.1007/s00198-006-0311-y. [DOI] [PubMed] [Google Scholar]
- 17.Gilissen C., Hoischen A., Brunner H.G., Veltman J.A. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andren L., Dymling J.F., Hogeman K.E., Wendeberg B. Osteopetrosis acro-osteolytica: a syndrome of osteopetrosis, acro-osteolysis and open sutures of the skull. Acta Chir Scand. 1962;124:496–507. [PubMed] [Google Scholar]
- 19.Maroteaux P., Lamy M. La pycnodysostose. Presse Med. 1962;70:999–1002. [PubMed] [Google Scholar]
- 20.Soliman A.T., Ramadan M.A., Sherif A., Aziz Bedair E.S., Rizk M.M. Pycnodysostosis: clinical, radiologic, and endocrine evaluation and linear growth after growth hormone therapy. Metabolism. 2001;50:905–911. doi: 10.1053/meta.2001.24924. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y., Cai T., Shi S., Wang W., Zhang Y., Mao T. Clinical and animal research findings in pycnodysostosis and gene mutations of cathepsin K from 1996 to 2011. Orphanet J Rare Dis. 2011;6:20. doi: 10.1186/1750-1172-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Hul W. Identification of genetic modifiers of monogenic (bone) diseases: new tools available, but with limitations. J Bone Miner Res. 2011;26:918–919. doi: 10.1002/jbmr.391. [DOI] [PubMed] [Google Scholar]
- 23.Rauch A., Wieczorek D., Graf E., Wieland T., Endele S., Schwarzmayr T. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 24.Need A.C., Shashi V., Hitomi Y., Schoch K., Shianna K.V., McDonald M.T. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sule G., Campeau P.M., Zhang V.W., Nagamani S.C., Dawson B.C., Grover M. Next-generation sequencing for disorders of low and high bone mineral density. Osteoporos Int. 2013;24:2253–2259. doi: 10.1007/s00198-013-2290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rehm H.L. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes associated to bone phenotypes.
Genotype of selected, putative modifier genes in our 6 CTSK-dependent patients.