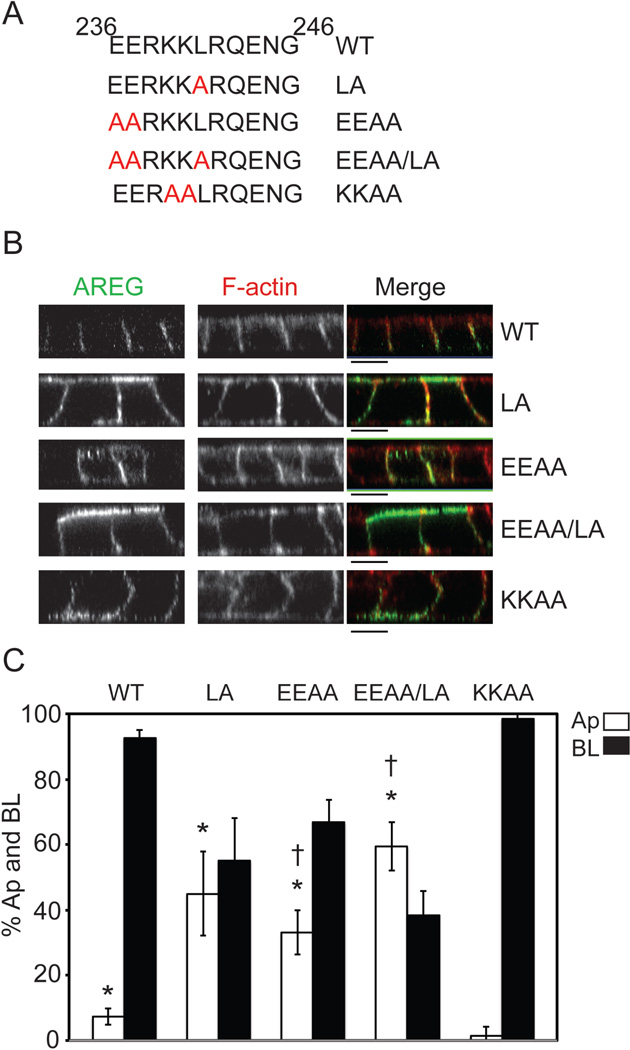
Figure 4. Amino acid substitutions within residues 236–246 identify a mono-leucine-based sorting signal.
(A) Amino acid composition of AREG cytoplasmic domain (ACD) between residues 236 and 246. Specific amino acid mutations are indicated in red. MDCK cells stably expressing the ACD mutants were generated and used to determine the BL sorting motif. Cells were polarized on Transwell filters and (B) surface stained for AREG (green) followed by permeabilization and F-actin (red) staining. Analysis was performed by confocal microscopy. Bars, 10 µm. (C) Polarized cells were selectively biotinylated on the Ap or BL surface, immunoprecipitated with anti-AREG mAb, separated by SDS-PAGE, transferred to nitrocellulose, and then probed with streptavidin-IRDye680. Results were scanned on an Odyssey scanner and quantitated using Odyssey software (n=3). Columns marked with an * are significantly different from WT. Columns marked with a † are significantly different from each other.