Figure 5.
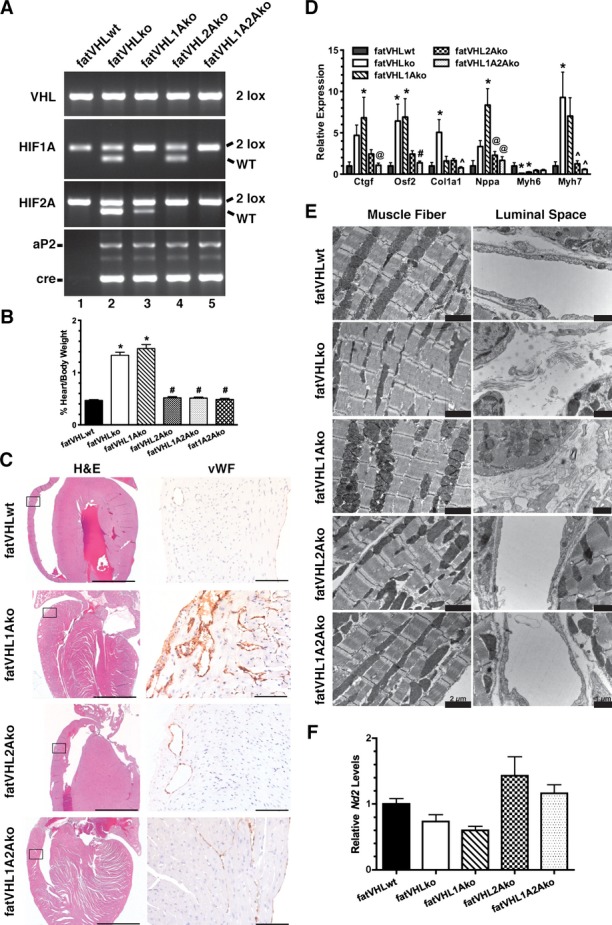
Hif2a deletion rescues the cardiac phenotype in fatVHLko mice. A, Generation of Vhl single KO (fatVHLko), Vhl/Hif1a double KO (fatVHL1Ako), Vhl/Hif2a double KO (fatVHL2Ako), and Vhl/Hif1a/Hif2a triple KO (fatVHL1A2Ako) mice, as shown by tail genotyping. Littermates without aP2‐cre were used as WT controls (fatVHLwt). B, Heart‐to‐body weight ratios (%). fatVHLwt: n=17; fatVHLko: n=9; fatVHL1Ako: n=6; fatVHL2Ako: n=5; fatVHL1A2Ako: n=7. P<0.0001 (1‐way ANOVA); *P<0.05 vs fatVHLwt; #P<0.05 vs fatVHLko or fatVHL1Ako (Dunnett's multiple‐comparison test). C, Histopathology of the heart indicates correction of cardiomegaly (H&E) and angiogenesis (anti‐vWF) in fatVHL2Ako and fatVHL1A2Ako, but not in fatVHL1Ako mice. Scales: 2000 μm (H&E) and 100 μm (anti‐vWF). D, Hif2a deletion reduces expression of hypertrophic cardiomyopathy‐associated genes in the heart, as shown by quantitative RT‐PCR. For each gene category, P<0.02 (1‐way ANOVA, fatVHLwt: n=6 to 7; fatVHLko: n=6 to 7; fatVHL1Ako: n=6 to 10; fatVHL2Ako: n=5; fatVHL1A2Ako: n=7 to 8). Significant differences (P<0.05) within each gene category: * vs WT, # vs fatVHLko or fatVHL1Ako, ^ vs fatVHLko. @ vs fatVHL1Ako (Dunnett's multiple‐comparison test). E, Electron microscopic analysis of left ventricular muscle shows increased fibrous deposits in fatVHLko and fatVHL1Ako hearts (right panel). F, Real‐time PCR quantification of the mitochondrial NADH dehydrogenase 2 (Nd2) in hearts of transgenic mice. Nd2 levels were normalized to the nucleus‐encoded DNA polymerase β. P=0.014 (1‐way ANOVA) with n=6 (fatVHLwt), 5 (fatVHLko), 6 (fatVHL1Ako), 7 (fatVHL2Ako), and 7 (fatVHL1A2Ako). HIF indicates hypoxia‐inducible factor; KO, knockout; VHL, von Hippel–Lindau; vWF, von Willebrand factor; WT, wild type.
