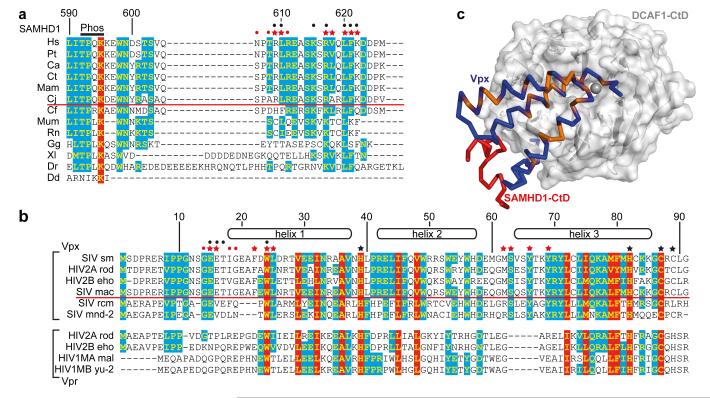
Figure 4. Species specificity of the SAMHD1-Vpx interaction.
(a) Sequence alignment of the C-termini of SAMHD1. 100% type-conserved residues are boxed red, 60% cyan. Residue numbers refer to hsSAMHD1-CtD. Red stars - side chains involved in Vpxsm-DCAF1-CtD binding, red dots - main chain interactions. Black dots - residues whose mutation impairs DDB1-DCAF1-Vpx binding 8. Species above the red line are HIV-2/SIV hosts, Phos - phosphorylation site. (b) Alignment of Vpx and Vpr proteins, coloured and annotated as in (a) numbering refers to Vpxsm. Red stars - side chains involved in SAMHD1-CtD binding, red dots - main chain interactions and black stars - zinc ligands. Black dots indicate residues whose mutation impairs SAMHD1 degradation. Proteins above the red line induce degradation of human SAMHD1, proteins below cannot. (c) Type-conserved amino acid residues in Vpxsm and VprHIV-1 (orange) mapped onto the Vpxsm (blue ribbon)-DCAF1-CtD (white surface) structure. SAMHD1-CtD is shown as red ribbon.