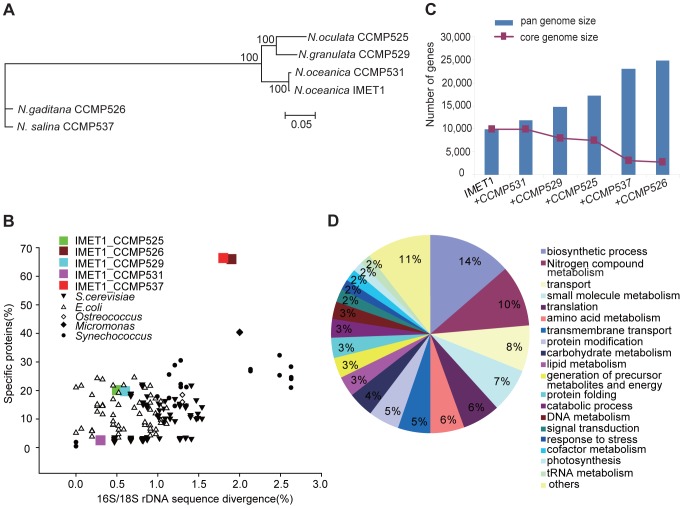
Figure 1. Structural features of the six Nannochloropsis genomes.
(A) Whole-genome based phylogeny of Nannochloropsis. A maximum likelihood (consensus) tree was generated using the PhyML program (JTT model) with 1,000 replicates based on the 1,085 six-way single-copy orthologous gene sets identified from the six Nannochloropsis genomes (Text S1). Percentages of replicate trees in the bootstrap test are shown next to the branches. (B) Genome divergence in Nannochloropsis. For each pair of genomes that consists of IMET1 and another Nannochloropsis strain, the percentages of strain-specific proteins versus their discrepancies in the full-length 18S rDNA sequence were plotted. Ten S. cerevisiae strains, eight E. coli strains, nine Synechococcus strains and two each of Ostreococcus and Micromonas were also included. For prokaryotic organisms including E. coli and Synechococcus, the percentages of strain-specific proteins were plotted against the discrepancies in 16S rDNA sequences. (C) The number of genes from the Nannochloropsis genomes and the Nannochloropsis core, with successive inclusion of each additional strain. (D) Functional categorization of Nannochloropsis core proteins. GO Slim terms corresponding to each GO term are presented.