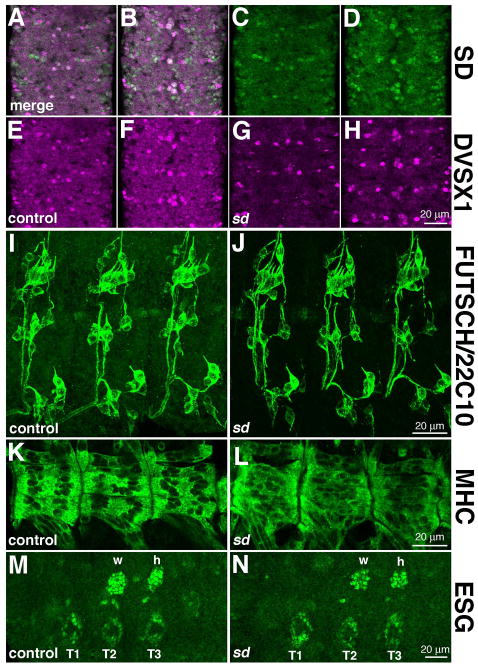
Figure 7. sd function is not required for global patterning of the PNS, CNS, muscles, and limb primordia.
(A–H) sd is not required for cells fate specification of the mVUMs and NB1-2 descendants in the ventral nerve cord. SD (green, A–D) is coexpressed with DVSX1 (magenta, A, B, E–H) in cells in the ventral nerve cord, including the mVUMs and NB1-2 descendants. The distribution of these cells is normal in Df(1)Exel6251 embryos, which are null for sd. (I, J) Antibody 22C10 was used to assess the distribution of cell bodies and axons in control (I) and sd3L embryos (J); these images are maximum projections of multiple confocal slices. We observed no reproducible difference between the wild type and mutant, showing that sd is not required for general PNS patterning. (K, L) Muscle structure was assessed using myosin heavy chain (MHC) in control (K) and Df(1)Exel6251 (L) embryos. Muscle structure is generally normally in sd mutant embryos, although altered distribution of nuclei was observed. (M, N) Distribution of ESG expressing cells in the dorsal and ventral limb primordia of control (M) and Df(1)Exel6251 (N) embryos. The normal distribution of these cells in sd mutant embryos confirms the observation that sd function is not required for the establishment of the limb primordia or the dorsal migration of the flight appendage cells (marked w, h) from the leg primordia (marked T1, T2, T3). Specimens are oriented with anterior up (A–H) or to the left (I–N).